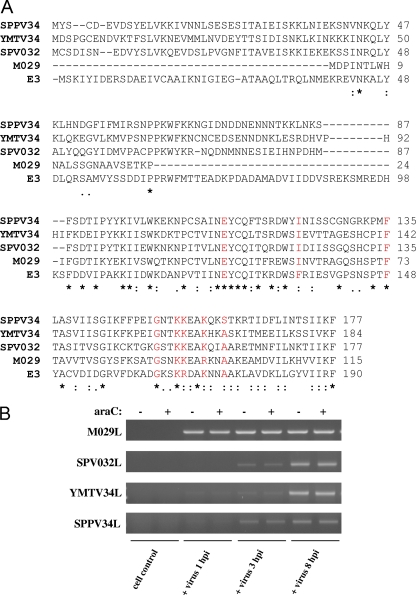
Fig. 1.
E3L orthologues are early genes whose protein products display significant amino acid divergence. (A) Protein sequence alignments were generated using the CLUSTAL W2 program. The “.” symbol denotes semiconserved substitutions, the “:” symbol denotes conserved substitutions, and the “*” symbol denotes complete conservation of the residue across all sequences. The amino acids highlighted in red indicate residues previously identified to contribute to E3 dsRNA binding. (B) OA3.Ts, CV-1, PK15, and RK13 cells were infected with SPPV, YMTV, SPV, and MYXV, respectively. Infections were performed in the presence or absence of araC, and cells were collected at 1, 3, and 8 hpi. The expression of SPPV34L, YMTV34L, SPV032L, and M029L was detected by RT-PCR in cells infected with the respective viruses.