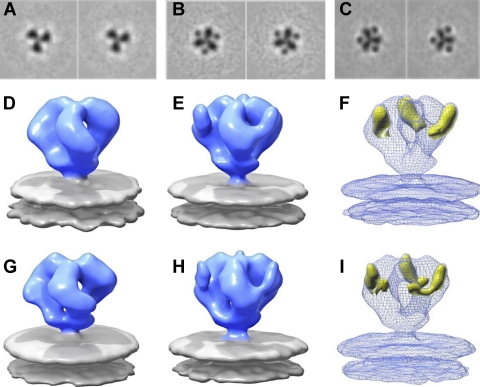
Fig. 2.
Comparison of trimeric SIV Env molecular architecture in native and sCD4-bound states. (A to C) Consecutive slices illustrating the density of the map at the apex of the spike in native SIVmneE11S Env (A), sCD4-bound SIVmneE11S Env (B), and sCD4-bound SIVmac239 Env (C). The densities from gp120 and sCD4 are clearly delineated. (D and E) Density map of SIVmneE11S Env displayed as an isosurface (blue and gray represent Env and viral membrane, respectively) in native (D) (58) and in sCD4-bound (E) conformations. (F) Superposition of density map from sCD4-Env complex on SIVmneE11S (shown in mesh) with positive difference density map (yellow isosurface) obtained by subtracting the native density map from the sCD4-bound density map. (G to I) Native, sCD4-bound, and superposition of liganded density maps with difference density maps for trimeric SIVmac239 Env, as in D to F for SIVmneE11S. Although both positive and difference densities were calculated for the respective maps, the positive difference density overwhelms the negative difference density peaks and are not visible at the threshold displayed. The mass contained within the displayed difference density threshold was estimated by procedures implemented in the EMAN software (39) and is ∼78 kDa, roughly matching the approximate molecular mass of 3 sCD4 molecules (24). No difference in structure was observed when sCD4 incubation was carried out at 37°C instead of 4°C.