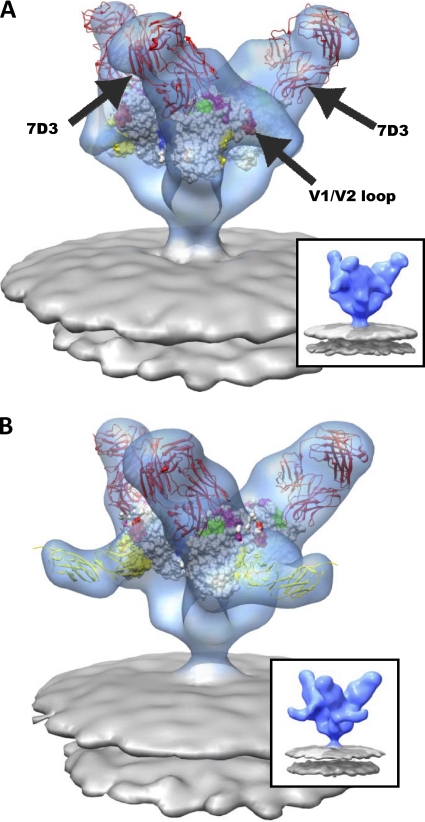
Fig. 5.
Molecular architecture of CCR5-binding site antibody (7D3) complexes on SIV CP-MAC Env with fitted gp120 coordinates. (A and B) Perspective views of 7D3-bound (red ribbons) SIV CP-MAC Env shown in blue transparent isosurface rendering in the absence (A) (58) and presence (B) of sCD4 (yellow ribbons). Insets show the same averaged maps as in the main figure panel, but without the fitted coordinates. Coordinate fits of a subset of gp120 X-ray coordinates from 1GC1 (A) or gp120 and sCD4 (B) rendered as in Fig. 3. Residues highlighted in magenta and blue indicate gp120 residues involved in coreceptor binding (47) and gp120 residues which have complete sequence conservation between HIV-1 and SIV, respectively. The 17b component of 1GC1 coordinates were used to model the 7D3 Fab fragment (red ribbons), since no crystal structure is available for 7D3 Fab, and it was then fitted into difference density calculated by subtracting the native SIV CP-MAC Env density from that of the antibody-bound map (see Materials and Methods for details).