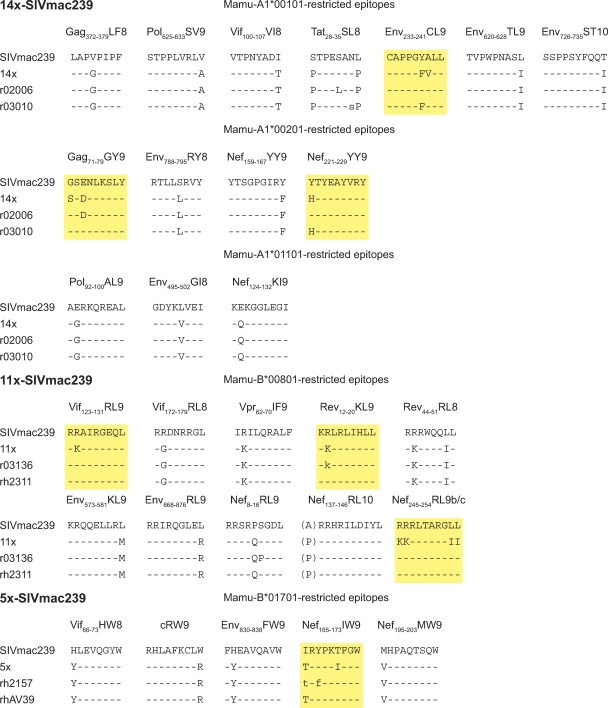
Fig. 3.
Plasma virus sequence. We selected two animals with high viral loads from each group for viral sequencing. Plasma virus samples from r02006, r03010, r03136, and rh2311 were sequenced approximately 26 weeks postinfection using whole-SIVmac239-genome Roche 454 sequencing (7). Mixed-base annotation indicates viral mutations encompassing more than 30% of total viral quasispecies. Capital-letter annotation indicates a mutation encompassing more than 70% of viral quasispecies. rh2157 and rhAV39 samples were sequenced using standard viral Sanger sequencing methods (22) at approximately 28 weeks postinfection. Reverting mutations are noted in yellow boxes. The Mamu-B*00801-restricted Nef245-254RL9 epitope encompasses two overlapping T cell epitopes (Table 1), both containing mutations that reverted. Dashes indicate residues identical to the wild-type sequence.