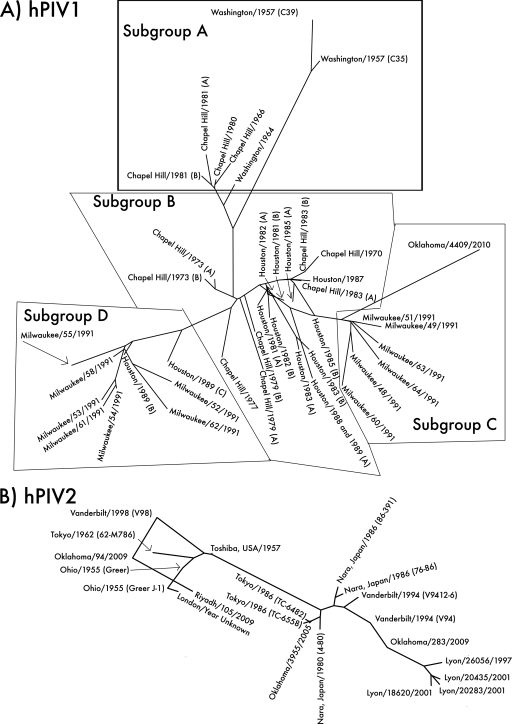
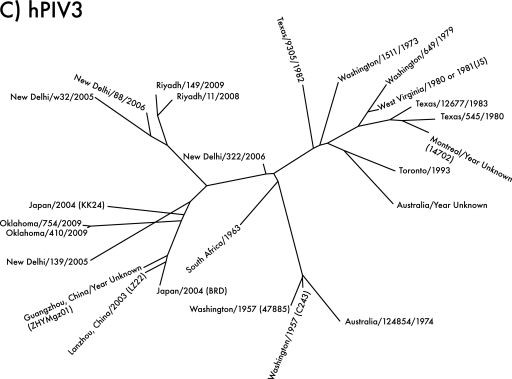
Fig. 2.
Unrooted phylogenetic trees of hPIV1, -2, and -3, based on sequences of type strains and local clinical isolates and clinical isolate sequences from GenBank. (A) Phylogeny of hPIV1. Local isolate Oklahoma/4409/2010 (far right) shares 93.7% amino acid sequence identity with the type strain Washington/1957 (C35) (top) and groups with the Milwaukee/1991 isolates previously identified as belonging to antigenic subgroup C (15). Assignment of sequences to subgroups is based on the Chapel Hill and Milwaukee isolates (14, 15). (B) Phylogeny of hPIV2. Local isolates Oklahoma/3955/2005, Oklahoma/94/2009, and Oklahoma/283/2009 are 94 to 96% identical in amino acid sequence to the type strain Ohio/1955 (Greer). Each Oklahoma isolate represents a different lineage, despite having originated in the same hospital over the course of only 4 years. hPIV2 strains tend to group geographically; note that the four Lyon isolates (lower right) group together, as do most of the isolates from Japan (center). However, closely related strains can also be isolated in different places at different times; for example, Ohio/1955, Tokyo/1962, Vanderbilt/1998, and Oklahoma/94/2009 share a branch of the tree. (C) Phylogeny of hPIV3. Local isolates Oklahoma/410/2009 and Oklahoma/754/2009 are 97 to 98% identical in amino acid sequence to the type strain Washington/1957 (C243). hPIV3 strains appear to group both temporally and geographically; strains from the New World and Australia before 1995 form one major branch of the tree (right), while strains from Asia after 1995 form the other (left). The Oklahoma/2009 isolates may be Asian in origin.