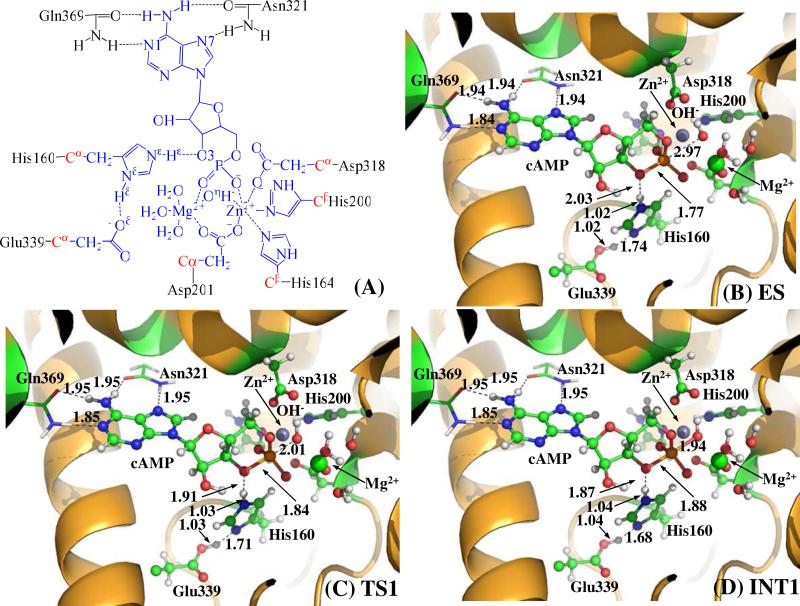
Figure 1.
(A) Division of the QM-MM systems for simulating the reaction stage 1 of PDE4-catalyzed cAMP hydrolysis. Atoms in blue are treated by the QM method. Six boundary carbon atoms (Cα, colored in red) are treated with the improved pseudobond parameters. All other atoms belong to the MM subsystem. (B to D) QM/MM-optimized geometries of key states of the reaction system for step 1, the nucleophilic attack of bridging hydroxide ion on the phosphorous atom of the cAMP. The geometries were optimized at the QM/MM(B3LYP/6-31G*:AMBER) level. The key distances in the figures are in angstrom. Carbon, oxygen, nitrogen and hydrogen atom are colored in greed, red, blue and white, respectively. The backbone of the protein is rendered in orange. The QM atoms are present as balls and sticks and the surrounding residues are rendered as sticks or lines.