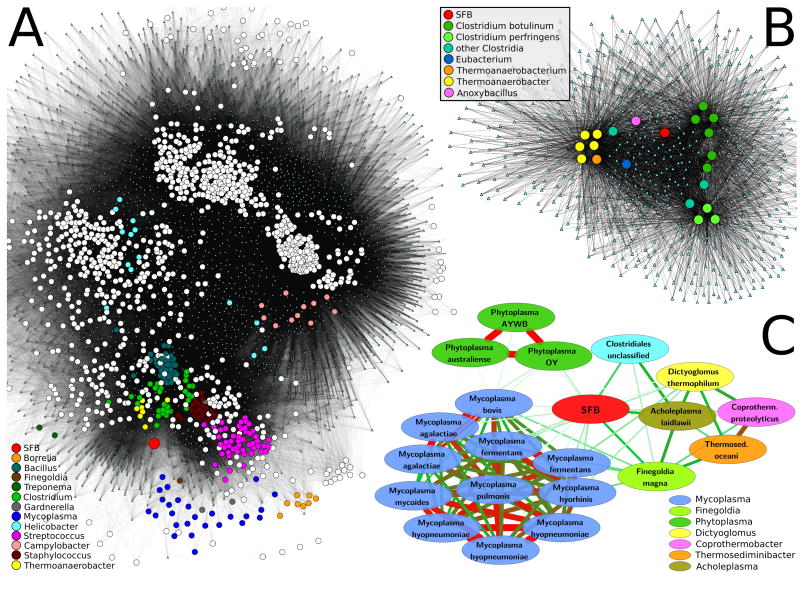
Figure 2. Genome-wide metabolic comparison between SFB and all sequenced genomes.
Analysis of microbial functional similarities based on shared orthologous gene families (A,B) and modules (C). A/B. The 1,209 genomes in KEGG and the 13,118 KEGG Orthology gene families (KOs) are reported as circles and small cyan triangles, respectively. Organisms are connected by edges to all gene families contained within their genome. A) Global network of all genomes for visual overview. SFB (large red circle) lies outside any cluster but is close to groups of several Firmicutes genera and in particular Clostridium, Thermoanaerobacter, Staphylococcus, and Streptococcus. Other genera, including Mycoplasma, Borrelia, Treponema, Finegoldia and Gardnerella are quantitatively similar to SFB (see text) but located in the network periphery due to overall reduced gene content. Despite their similarity to SFB in terms of genome size and host environment, Helicobacter and Campylobacter are located in different regions of the network suggesting different functional specialization. B) Sub-network of SFB and the 20 most similar organisms (Tversky index 0.75, Table 2); KOs included in at least two organisms are depicted. SFB is functionally distinct from both the cluster of Clostridia (further differentiated as C. botulinum, C. perfringens, and “other”) and the Thermoanaerobacteria. C) Metabolic comparison based on functional genomic potential (Table 2) highlights SFB’s similarity to several Myco/Phyto/Acholeplasma and to Finegoldia magna (for which only one genome is available). The width and color (white = low, green = medium, red = high) of edges reflect relationship strength. Again, despite considering only the most functionally similar organisms, SFB is not directly included in the clusters they form.