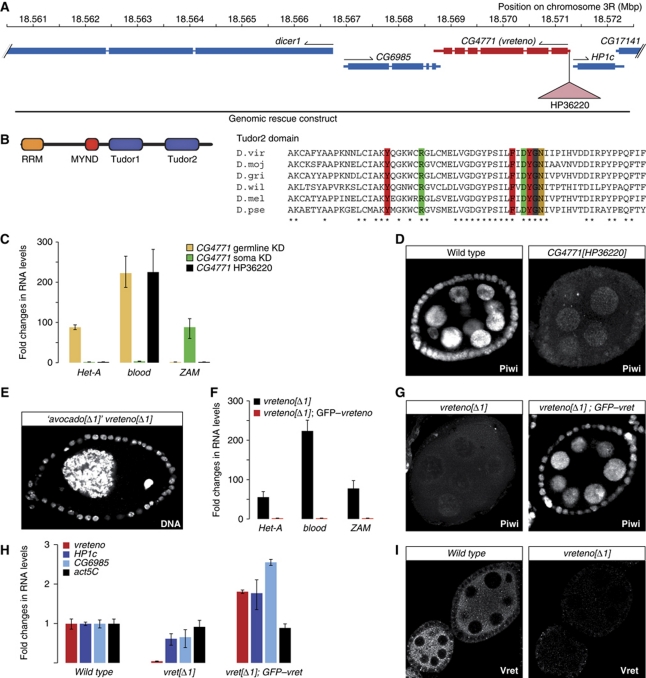
Figure 3.
Vreteno is a novel piRNA pathway member. (A) Overview of the CG4771 (vreteno) genomic locus indicating flanking genes (blue), the HP36220-insertion site (pink triangle) and the extent of the genomic rescue construct. (B) Cartoon of the CG4771 protein domain structure and sequence alignment of the C-terminal TUDOR domain in distantly related Drosophilids (virilis, mojavensis, grimshawi, willistoni, melanogaster, pseudoobscura). Aromatic cage residues and the conserved Arg/Asp residues colour coded as in Figure 1B. (C) Changes in steady-state transposon levels (n=3; s.d.) upon CG4771 knockdown (normalized to no-hairpin controls) in soma (green) or germline (beige) in comparison to those in CG4771[HP36220] mutants (black; normalized to heterozygotes). (D) Immunostaining of Piwi in wild-type and CG4771[HP36220] mutant egg chambers. (E) The occasionally observed egg chamber morphology of CG4771[Δ1] (vreteno) mutants, which originally led us to name the gene ‘avocado’ (DNA stained with DAPI). (F) RNA levels of CG4771, of the flanking genes HP1c and CG6985 and of actin-5C in vreteno[Δ1] mutant ovaries compared with vreteno[Δ1]; GFP–vreteno rescued ovaries (values normalized to w[1118] controls). (G) Immunostaining of Piwi in vreteno[Δ1] mutant egg chambers and in vreteno[Δ1] mutant egg chambers expressing a GFP–vreteno rescue construct. (H) Steady-state RNA levels of the HeT-A, blood and ZAM transposons in vreteno[Δ1] mutant ovaries compared with vreteno[Δ1]; EGFP–vreteno rescued ovaries (values normalized to heterozygous siblings; n=3; error bars indicate s.d.). (I) Immunostaining of Vreteno in wild-type and vreteno[Δ1] mutant egg chambers at identical microscope settings.