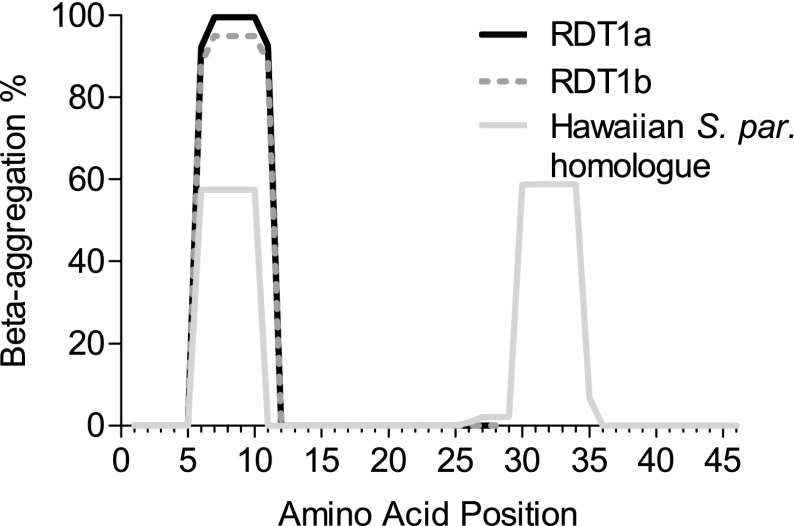
FIG. 5.–
Protein aggregation propensity (%) along the length of the putative protein, as predicted by TANGO. The y axis indicates the estimated likelihood that a peptide would be found in an aggregated structure rather than, for example, as an α-helix or β-sheet (Fernandez-Escamilla et al. 2004). These probabilities are based on thermodynamic stability and the Boltzmann equation and apply to the peptide in isolation from the rest of the protein. Both Saccharomyces cerevisiae RDT1 alleles have a predicted aggregation-prone sequence from amino acids 6–11 inclusive. No aggregation is predicted for S. mikatae, whose amino acids are not homologous in this region due to a frameshift. The single strain of S. paradoxus that contained an ORF shows a weak aggregation propensity, at a position shifted by one amino acid.