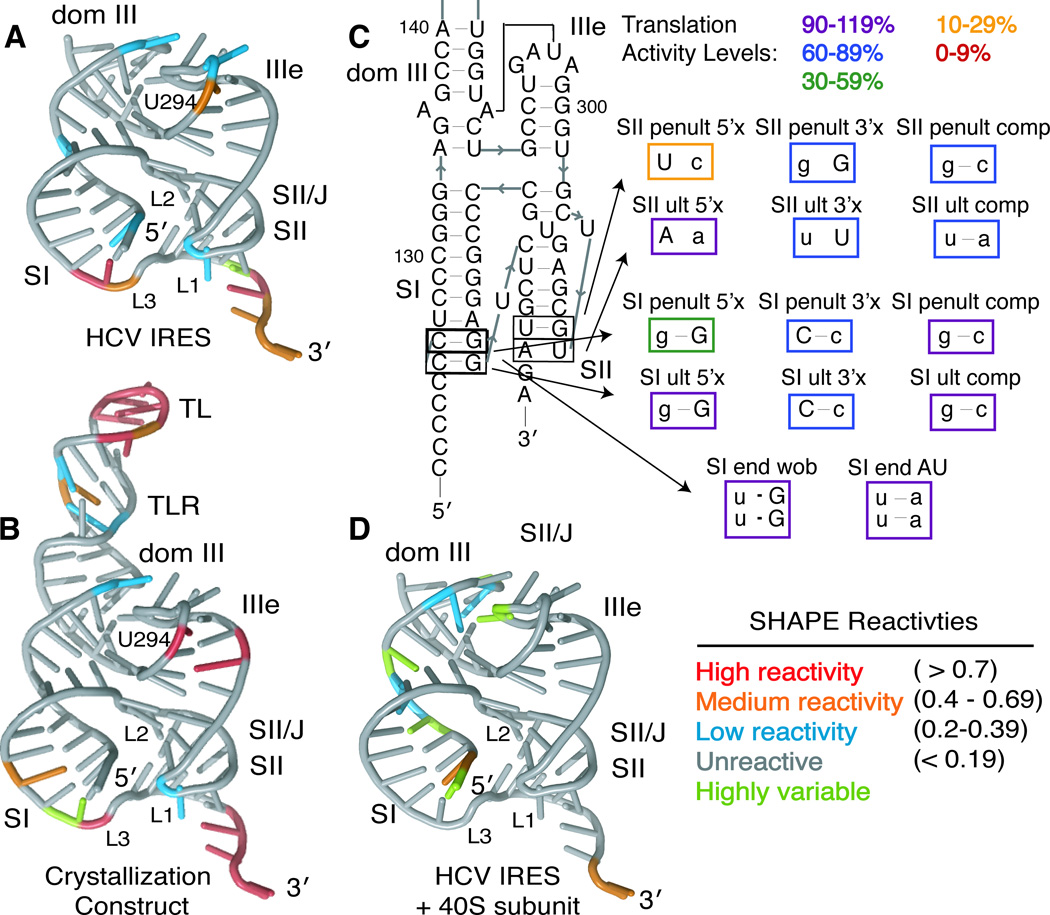
Figure 4. Solution structural probing and pseudoknot-domain dynamics.
SHAPE modification reactivities of individual nucleotides from the (A) full-length HCV IRES or (B) the crystallization construct are mapped onto the crystal structure, colored according to reactivity (see legend). (C) Mutational analysis of the ultimate and penultimate predicted base pairs of SI and SII. Mutated nucleotides are shown in lowercase and mutants are boxed in colors according to their translation activity, relative to WT, as defined in the legend. (D) SHAPE modification reactivities of individual nucleotides from the full-length HCV IRES in the presence of 40S ribosomal subunits are shown on the crystal structure, colored according to reactivity (see legend). See also Figure S2.