Abstract
MicroRNAs (miRNAs) are 22 nt non-coding RNAs that regulate expression of downstream targets by messenger RNA (mRNA) destabilization and translational inhibition. A large number of eukaryotic mRNAs are targeted by miRNAs, with many individual mRNAs being targeted by multiple miRNAs. Further, a single miRNA can target hundreds of mRNAs, making these small RNAs powerful regulators of cell fate decisions. Such regulation by miRNAs has been observed in the maintenance of the embryonic stem cell (ESC) cell cycle and during ESC differentiation. MiRNAs can also promote the dedifferentiation of somatic cells to induced pluripotent stem cells. During this process they target multiple downstream genes, which represent important nodes of key cellular processes. Here, we review these findings and discuss how miRNAs may be used as tools to discover novel pathways that are involved in cell fate transitions using dedifferentiation of somatic stem cells to induced pluripotent stem cells as a case study.
MicroRNA-mediated suppression of mRNAs
Details of how miRNAs recognize and downregulate their downstream mRNA targets can be found in other excellent reviews [1–3] and the topic is only briefly discussed here. MiRNAs are approximately 22nt long small RNAs that regulate their targets through incomplete nucleotide complementation. Most miRNA-mRNA targeting occurs through base-pairing between a short sequence located at the 5′ end of the miRNA, called the seed sequence, and its mRNA target. This seed sequence, ranging from nucleotide positions 2–8 in the miRNA, largely defines the miRNA’s downstream targets and hence is the basis of most target prediction programs (reviewed in [1]). Exceptions to seed sequence pairing exist, but these make up a much smaller repertoire of miRNA-mRNA targeting events [4]. The exact consequence of miRNA-mRNA pairing is controversial, although the end result is both a decrease in mRNA and protein levels [5–8]. Interestingly, within cells, pairing between miRNA-mRNA can be regulated by various mechanisms including co-expression of the target and miRNA, alternative poly-adenylation leading to alternative 3′UTRs of mRNAs, and protein based enhancement or suppression of specific miRNA-mRNA pairing [9–17]. Ultimately, this minimal requirement of nucleotide complementation for miRNA-mRNA pairing results in a single miRNA suppressing hundreds of targets [1].
MicroRNA Redundancy
Studies in miRNA function have been complicated not only by the fact that a single miRNA regulates multiple targets, but also by functional redundancy among miRNAs in many, if not most, biological processes [18–20]. This redundancy results in part from miRNAs existing in large families sharing common seed sequences that can be co-expressed in the same cell, and hence share overlapping downstream mRNA targets [20]. Redundancy also occurs at the level of co-targeting where multiple distinct miRNAs with very different sequences commonly target a single transcript through non-overlapping sites [21].
A powerful means of overcoming this redundancy in order to study individual miRNA function in a given biological setting is to first remove all miRNAs and then reintroduce individual miRNAs mimics. Global removal of miRNAs is made possible by deleting genes encoding proteins responsible for the processing of miRNAs. The biogenesis of most miRNAs requires two essential processing steps: primary- to precursor-miRNA by the DGCR8/DROSHA complex and precursor- to mature miRNA by DICER [22,23]. Knockout alleles for all three genes encoding these proteins have been made, thus providing powerful tools with which the function of individual miRNAs can be studied [24–28].
MicroRNA functions in embryonic stem cells
Deletion of Dgcr8 or Dicer in embryonic stem cells (ESCs) results in two interesting phenotypes, a proliferation defect and a block in differentiation [20,26–29]. The proliferation defect is associated with an accumulation of cells in the G1 phase of the cell cycle. In a screen conducted to identify miRNAs that could rescue this phenotype, members of the miR-290 and 302 clusters were uncovered [20]. The miR-290 cluster is highly expressed in mouse ES cells, while the 302 cluster is highly expressed in human ES cells [30,31]. The specific members of these clusters that rescue proliferation share a common seed sequence and are collectively termed the ESCC family of miRNAs for ESC cell cycle promoting miRNAs. The ESCC miRNAs target a number of important cell cycle regulators. These included the CDK inhibitor Cdkn1a, the tumor suppressor Lats2, and members of the RB family of proteins. Expression of Cdkn1a without its 3′UTR in wild-type ES cells partially recapitulates the cell cycle phenotype of Dgcr8 knockout cells, indicating that Cdkn1a can only partially explain the effect of ESCC miRNA loss [20].
In addition to the cell cycle defect, Dgcr8 knockout ES cells fail to downregulate pluripotency factors when cultured under differentiation-inducing conditions [28,29]. Introduction of members of the let-7 family of miRNAs can rescue this defect. Let-7 miRNAs are highly expressed in differentiated tissues and hence are well positioned to repress the self-renewal program [32]. Profiling after introducing let-7 into Dgcr8 knockout cells combined with bioinformatic analyses reveals a large number of likely targets for this family of miRNAs [29]. These targets include multiple well-known pluripotency genes such as nMyc, Sall4, and Lin28. Interestingly, Lin28 is an inhibitor of let-7 biogenesis producing a double negative feedback loop [33]. Members of the let-7 family of miRNAs are not the only miRNAs shown to promote silencing of ESC self-renewal. For example, miR-145 and miR-134 silence self-renewal of human and mouse ESCs respectively. MiR-145 targets Oct4, Sox2 and Klf4, while miR-134 targets Nanog and LRH1, all important pluripotency genes [34,35].
Collectively these studies show that miRNAs are critical regulators of the switch between ESC self-renewal and differentiation. While a single miRNA family, the ESCC family, seems to be largely responsible for promoting ESC self-renewal, multiple miRNAs can promote differentiation [36]. Importantly, each of these miRNAs is predicted to have many downstream mRNA targets.
One microRNA – many targets
Historically, most miRNA phenotypic studies have focused on one or a small number of targets. These include classic examples such as the regulation of lin-14 by the small RNA lin-4 and the regulation of lin-41 by let-7 in worms [37,38]. However, it is becoming increasingly apparent that regulation of a single target by a miRNA is unlikely to explain how they function in most situations. Indeed, the power of miRNAs influencing a cell fate decision is likely through their ability to alter levels of many genes and pathways simultaneously. For example, the introduction of a lineage-specific miRNA (miR-124) into HeLa cells (tumor cells of cervical origin) results in a global shift in their expression profile towards that of a cell of the neural lineage. Much of the shift can be attributed to direct targeting by the miRNA, as there is a strong enrichment of seed matches in the 3′UTRs of downregulated genes [7].
Functional experiments have corroborated these early findings. For example, a number of miR-124’s targets have since been confirmed as direct targets with functional relevance to neuronal differentiation [39–41]. Another example is miR-31, which is repressed in metastatic breast cancer cells, while its targets including RhoA, Fzd3, ITGA5, M-RIP, MMP16 and RDX are upregulated [42]. Co-expression of three targets, ITGA5, RDX and Rho in breast cancer cells overcomes the block in metastasis upon overexpression of miR-31, while individual targets had only a partial effect [43]. Additional examples of miRNAs with multiple targets in common processes are provided in Table 1. However, this list is not comprehensive.
Table 1. Individual miRNAs regulate the expression of multiple targets.
Listed are a few examples of miRNAs and some select targets that are involved in cell fate transitions.
| MicroRNA | Validated Targets | Context | Organism | Reference |
|---|---|---|---|---|
| miR-124 | NeuroD1, Baf53a, Ptbp1, Scp1, Sox9, Dlx2, Jag1 | Neural development | Xenopus, Mouse, Chick | [39–41,66,67] |
| miR-31 | RhoA, Fzd3, Itga5, Mmp16, M-Rip, Rdx | Metastasis | Human | [42] |
| miR-19 | Pten, Ppp2r5e, Prkaa1, Bim | Leukemia | Mouse | [68] |
| miR-21 | Tiam1, Anp32a, Smarca4, P12 Cdk2ap1 | Cancer | Human | [69–71] |
| miR-200 | Zeb1, Zeb2, Ets1, Suz12, Bmi1 | Cancer | Human and mouse | [72–78] |
| miR-294, miR-302, miR-372 | Cdkn1a, Rbl2, Lats2, Tgfbr2, RhoC, Mecp1–p66, Mecp2, Aof1, Aof2, Cyclin D1 | Embryonic stem cells and dedifferentiation | Mouse and human | [20,62,63,65,79–83] |
| Let-7 | Hmga2, Ras, cMyc, NMyc, Sall4, Lin28, Trim71, Cyclin D1, Tlx | Differentiation and cancer | Mouse, human and C. elegans | [29,33,84–89] |
Induced pluripotency – resetting the clock
In 2006, Yamanaka and colleagues demonstrated that the introduction of four transcription factors, Oct3/4, Sox2, Klf4 and cMyc into somatic cells, resulted in their conversion into pluripotent cells, called induced pluripotent stem cells [44]. While many groups thereafter have also succeeded in creating induced pluripotent stem cells from a variety of starting cell populations, little is known about all the changes that a cell has to undergo to become pluripotent. Furthermore, reprogramming remains to date a slow and relatively inefficient process and a better understanding of downstream events after introduction of the Yamanaka factors would enable us to develop patient-specific iPS lines in a faster and safer manner. Often cells become trapped in a partially reprogrammed state following introduction of the factors [45–47]. These cells are self-renewing, but not pluripotent. The comparison of partially reprogrammed cells to fully reprogrammed cells or embryonic stem cells or to the starting population of differentiated cells has provided valuable insight into the roadblocks during the process of reprogramming [46,47]. Such comparisons have revealed that reprogramming of MEFs to iPS cells requires a genome-wide alteration of epigenetic marks. This includes the conversion of monovalent histone methylation marks such as H3K4me3 or H3K27me3 to bivalent marks at developmental genes; the reactivation of transcription of pluripotency genes and the loss of DNA methylation from a large part of the genome [48]. In support of the role of epigenetic modifications during dedifferentiation, treatment of cells with small molecule inhibitors to HDACs, DNA methyltransferases and the G9a methyltranferase have been shown to enhance the process of reprogramming [46,49–51]. Transcriptional profiling of partially reprogrammed cells indicates that while a number of genes associated with cell proliferation and DNA synthesis are upregulated, genes such as Cdkn1a which regulate cell cycle checkpoints are also expressed at high levels. Indeed, a number of studies have reported that knockdown of Cdkn1a, p53 or Ink4/Arf enhance the efficiency of reprogramming [52–57]. In addition to the involvement of epigenetics and cell cycle in the process of reprogramming, the conversion of cells from a mesenchymal to an epithelial state is also required [58,59]. Thus, the acquisition of pluripotency is associated with a number of changes in the state of the cell, of which only a few are known.
MicroRNAs and induced pluripotency: pathway discovery during cell fate transitions
Similar to small molecules, miRNAs can influence reprogramming. The ESCC miRNAs and the closely related miR-106 family enhance the efficiency of reprogramming [60–63]. Indeed, it was recently reported that the miR-302 cluster (consisting of miRs-302a–d and miR-367) alone could produce iPSCs from both mouse and human fibroblasts [64]. Interestingly, the promoters of the miR-290 cluster (hsa-miR-371/372/373 in human) and miR-302 clusters are bound and regulated by the original Yamanaka factors, Oct4, Sox2, and cMyc [32,60,65]. Therefore, these transcription factors are likely at least in part acting through these miRNAs to promote de-differentiation of somatic cells.
These results lead to the obvious question of what downstream targets underlie the miRNA’s remarkable capacity to revert an adult cell back to an embryonic stem cell. Hints come from profiling experiments following the introduction of the ESCC miRNAs into Dgcr8 knockout cells [29]. Similar to the miR-124 experiments in HeLa cells described above, hundreds of transcripts with seed matches in their 3′UTR are downregulated. Interestingly, there is a highly significant enrichment of seed matches in the open reading frame and 3′UTR, consistent with targeting in both regions of the transcript. In order to gain a better understanding of pathways regulated by these miRNAs during reprogramming, a subset of these targets have been further characterized [63]. This subset was selected on the basis of their known role in potentially relevant cellular processes including cell cycle regulation, epithelial-mesenchymal transition (EMT), vesicular trafficking, cell signaling and epigenetic modifications. Knockdown of any single individual target within this subset of targets only marginally increases the efficiency of reprogramming, much less so than the miRNA itself. This finding suggests that these targets/cellular processes work together to promote reprogramming. Indeed, co-suppression of two targets further enhances reprogramming. Therefore, these miRNAs uncovered cellular pathways, whose role in reprogramming was not fully appreciated and that act synergistically to promote the induction of pluripotency.
Using miRNAs as tools to dissect mechanisms of cell fate transitions
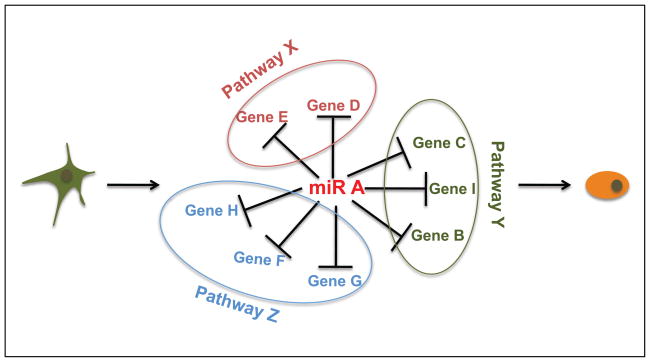
While the promiscuous nature of miRNA targeting defies simplification of their roles, we propose that they can be used as a powerful tool to uncover the multiple pathways/cellular processes that underlie cell fate decisions (Fig. 1). The approach takes advantage of the fact that the hundreds of mRNA targets have been evolutionarily selected to have a defined cellular outcome. That is, the identification of miRNAs that promote a specific outcome, combined with the identification and functional characterization of all its targets can provide a holistic picture of the events that must occur. As mentioned in this review, a number of studies performed on a smaller scale have already provided evidence for the success of such an approach. However, a full-scale study functionally testing all downstream targets has yet to be performed.
Figure 1. Pathway discovery using microRNAs.
Introduction of single miRNAs that regulate cell fate decisions followed by profiling can uncover numerous target genes that are key nodes of signaling pathways.
Acknowledgments
This work was supported by funds to RB from Leona M. and Harry B. Helmsley Charitable Trust, NIH (K08 NS48118, R01 NS057221, and U54HD055764 pilot), California Institute of Regenerative Medicine (CIRM) (Seed Grant RS1-00161, New Faculty Award RN2-00906), the American Health Assistance Foundation (formerly Stem Cell Research Foundation) and the Pew Charitable Trust. We thank members of the Blelloch lab for their helpful comments and for critically reading the manuscript.
References
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
- 1.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Djuranovic S, Nahvi A, Green R. A parsimonious model for gene regulation by miRNAs. Science. 2011;331:550–553. doi: 10.1126/science.1191138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 4.Shin C, Nam JW, Farh KK, Chiang HR, Shkumatava A, Bartel DP. Expanding the microRNA targeting code: functional sites with centered pairing. Mol Cell. 2010;38:789–802. doi: 10.1016/j.molcel.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6*.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. The authors perform ribosome-footprinting after introduction of a miRNA and conclude that miRNA-mediated repression is mostly attributed to destabilization of the mRNA, rather than translational repression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 8.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 9.Hammell CM, Lubin I, Boag PR, Blackwell TK, Ambros V. nhl-2 Modulates microRNA activity in Caenorhabditis elegans. Cell. 2009;136:926–938. doi: 10.1016/j.cell.2009.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kedde M, Strasser MJ, Boldajipour B, Oude Vrielink JA, Slanchev K, le Sage C, Nagel R, Voorhoeve PM, van Duijse J, Orom UA, et al. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell. 2007;131:1273–1286. doi: 10.1016/j.cell.2007.11.034. [DOI] [PubMed] [Google Scholar]
- 11.Kedde M, van Kouwenhove M, Zwart W, Oude Vrielink JA, Elkon R, Agami R. A Pumilio-induced RNA structure switch in p27-3′ UTR controls miR-221 and miR-222 accessibility. Nat Cell Biol. 2010;12:1014–1020. doi: 10.1038/ncb2105. [DOI] [PubMed] [Google Scholar]
- 12.Liu H, Kohane IS. Tissue and process specific microRNA-mRNA co-expression in mammalian development and malignancy. PLoS One. 2009;4:e5436. doi: 10.1371/journal.pone.0005436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13*.Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. This study shows that messenger RNAs with shorter 3′ untranslated regions are frequently observed in cancer cells due to alternative cleavage and polyadenylation. These shorter isoforms display greater stability due to loss of miRNA binding sites in their 3′UTRs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwamborn JC, Berezikov E, Knoblich JA. The TRIM-NHL protein TRIM32 activates microRNAs and prevents self-renewal in mouse neural progenitors. Cell. 2009;136:913–925. doi: 10.1016/j.cell.2008.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takeda Y, Mishima Y, Fujiwara T, Sakamoto H, Inoue K. DAZL relieves miRNA-mediated repression of germline mRNAs by controlling poly(A) tail length in zebrafish. PLoS One. 2009;4:e7513. doi: 10.1371/journal.pone.0007513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tian B, Hu J, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18*.Alvarez-Saavedra E, Horvitz HR. Many families of C. elegans microRNAs are not essential for development or viability. Curr Biol. 2010;20:367–373. doi: 10.1016/j.cub.2009.12.051. This study tests phenotypes resulting from deletion of entire families of miRNAs in C. elegans. Surprisingly, for many families, there is no phenotype. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miska EA, Alvarez-Saavedra E, Abbott AL, Lau NC, Hellman AB, McGonagle SM, Bartel DP, Ambros VR, Horvitz HR. Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS Genet. 2007;3:e215. doi: 10.1371/journal.pgen.0030215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, Baskerville S, Shenoy A, Babiarz JE, Baehner L, Blelloch R. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat Genet. 2008;40:1478–1483. doi: 10.1038/ng.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21*.Tsang JS, Ebert MS, van Oudenaarden A. Genome-wide dissection of microRNA functions and cotargeting networks using gene set signatures. Mol Cell. 2010;38:140–153. doi: 10.1016/j.molcel.2010.03.007. The authors develop a computational method, mirBridge that groups miRNA targets into networks. They also identify wide-spread co-targeting where a single mRNA can be targeted by multiple miRNAs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Babiarz JE, Blelloch R. Small RNAs - their biogenesis, regulation and function in embryonic stem cells. 2008. [PubMed] [Google Scholar]
- 23.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 24.Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 25.Chong MM, Rasmussen JP, Rudensky AY, Littman DR. The RNAseIII enzyme Drosha is critical in T cells for preventing lethal inflammatory disease. J Exp Med. 2008;205:2005–2017. doi: 10.1084/jem.20081219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci U S A. 2005;102:12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29*.Melton C, Judson RL, Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature. 2010;463:621–626. doi: 10.1038/nature08725. This study shows opposing roles for the ESCC and let-7 family of miRNAs in the switch between ESC self-renewal and differentiation. Furthermore, it characterizes the downstream pathways that underly their antagonistic roles. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific MicroRNAs. Dev Cell. 2003;5:351–358. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- 31.Suh MR, Lee Y, Kim JY, Kim SK, Moon SH, Lee JY, Cha KY, Chung HM, Yoon HS, Moon SY, et al. Human embryonic stem cells express a unique set of microRNAs. Dev Biol. 2004;270:488–498. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 32.Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, Wulczyn FG. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol. 2008;10:987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 34.Tay YM, Tam WL, Ang YS, Gaughwin PM, Yang H, Wang W, Liu R, George J, Ng HH, Perera RJ, et al. MicroRNA-134 modulates the differentiation of mouse embryonic stem cells, where it causes post-transcriptional attenuation of Nanog and LRH1. Stem Cells. 2008;26:17–29. doi: 10.1634/stemcells.2007-0295. [DOI] [PubMed] [Google Scholar]
- 35*.Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137:647–658. doi: 10.1016/j.cell.2009.02.038. This study shows that miR-145 is upregulated and directly targets pluripotent factors such as Oct4, Sox2 and Klf4 during the differentiation of human embryonic stem cells. [DOI] [PubMed] [Google Scholar]
- 36.Melton C, Blelloch R. MicroRNA Regulation of Embryonic Stem Cell Self-Renewal and Differentiation. Adv Exp Med Biol. 2010;695:105–117. doi: 10.1007/978-1-4419-7037-4_8. [DOI] [PubMed] [Google Scholar]
- 37.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 38.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 39.Visvanathan J, Lee S, Lee B, Lee JW, Lee SK. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 2007;21:744–749. doi: 10.1101/gad.1519107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoo AS, Staahl BT, Chen L, Crabtree GR. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature. 2009;460:642–646. doi: 10.1038/nature08139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42*.Valastyan S, Reinhardt F, Benaich N, Calogrias D, Szasz AM, Wang ZC, Brock JE, Richardson AL, Weinberg RA. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell. 2009;137:1032–1046. doi: 10.1016/j.cell.2009.03.047. Authors show that miR-31 expression is repressed in metastatic breast cancer cells. Overexpression of individual targets of this miRNA fails to recapitulate the entire process of metastasis, indicating that multiple targets cooperate to mediate this process. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Valastyan S, Benaich N, Chang A, Reinhardt F, Weinberg RA. Concomitant suppression of three target genes can explain the impact of a microRNA on metastasis. Genes Dev. 2009;23:2592–2597. doi: 10.1101/gad.1832709. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 44.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 45.Lowry WE, Richter L, Yachechko R, Pyle AD, Tchieu J, Sridharan R, Clark AT, Plath K. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proc Natl Acad Sci U S A. 2008;105:2883–2888. doi: 10.1073/pnas.0711983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sridharan R, Tchieu J, Mason MJ, Yachechko R, Kuoy E, Horvath S, Zhou Q, Plath K. Role of the murine reprogramming factors in the induction of pluripotency. Cell. 2009;136:364–377. doi: 10.1016/j.cell.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Plath K, Lowry WE. Progress in understanding reprogramming to the induced pluripotent state. Nat Rev Genet. 2011;12:253–265. doi: 10.1038/nrg2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huangfu D, Osafune K, Maehr R, Guo W, Eijkelenboom A, Chen S, Muhlestein W, Melton DA. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat Biotechnol. 2008;26:1269–1275. doi: 10.1038/nbt.1502. [DOI] [PubMed] [Google Scholar]
- 50.Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, Chen AE, Melton DA. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol. 2008;26:795–797. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi Y, Desponts C, Do JT, Hahm HS, Scholer HR, Ding S. Induction of pluripotent stem cells from mouse embryonic fibroblasts by Oct4 and Klf4 with small-molecule compounds. Cell Stem Cell. 2008;3:568–574. doi: 10.1016/j.stem.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 52.Banito A, Rashid ST, Acosta JC, Li S, Pereira CF, Geti I, Pinho S, Silva JC, Azuara V, Walsh M, et al. Senescence impairs successful reprogramming to pluripotent stem cells. Genes Dev. 2009;23:2134–2139. doi: 10.1101/gad.1811609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Utikal J, Polo JM, Stadtfeld M, Maherali N, Kulalert W, Walsh RM, Khalil A, Rheinwald JG, Hochedlinger K. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009;460:1145–1148. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marion RM, Strati K, Li H, Murga M, Blanco R, Ortega S, Fernandez-Capetillo O, Serrano M, Blasco MA. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li H, Collado M, Villasante A, Strati K, Ortega S, Canamero M, Blasco MA, Serrano M. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460:1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, Wahl GM, Belmonte JC. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460:1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao Y, Yin X, Qin H, Zhu F, Liu H, Yang W, Zhang Q, Xiang C, Hou P, Song Z, et al. Two supporting factors greatly improve the efficiency of human iPSC generation. Cell Stem Cell. 2008;3:475–479. doi: 10.1016/j.stem.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 58*.Li R, Liang J, Ni S, Zhou T, Qing X, Li H, He W, Chen J, Li F, Zhuang Q, et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. Authors show that the induction of epithelial genes by the Yamanaka factors is an important step during the process of reprogramming. Blocking this step can result in a failure to reprogram cells. [DOI] [PubMed] [Google Scholar]
- 59*.Samavarchi-Tehrani P, Golipour A, David L, Sung HK, Beyer TA, Datti A, Woltjen K, Nagy A, Wrana JL. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7:64–77. doi: 10.1016/j.stem.2010.04.015. The authors profile cells undergoing reprogramming at different time points and determine that the induction of genes regulating MET is a key step in this process. [DOI] [PubMed] [Google Scholar]
- 60*.Judson RL, Babiarz JE, Venere M, Blelloch R. Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat Biotechnol. 2009;27:459–461. doi: 10.1038/nbt.1535. The authors describe for the first time that miRNAs highly expressed in ES cells can enhance the efficiency of reprogramming mouse embryonic fibroblasts to induced pluripotent stem cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li Z, Yang CS, Nakashima K, Rana TM. Small RNA-mediated regulation of iPS cell generation. EMBO J. 2011;30:823–834. doi: 10.1038/emboj.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liao B, Bao X, Liu L, Feng S, Zovoilis A, Liu W, Xue Y, Cai J, Guo X, Qin B, et al. Microrna cluster 302–367 enhances somatic cell reprogramming by accelerating a mesenchymal-to-epithelial transition. J Biol Chem. 2011 doi: 10.1074/jbc.C111.235960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63*.Subramanyam D, Lamouille S, Judson RL, Liu JY, Bucay N, Derynck R, Blelloch R. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat Biotechnol. 2011 doi: 10.1038/nbt.1862. Authors show that the ESCC miRNAs miR-302b and miR-372 act through the regulation of multiple targets within defined pathways to promote dedifferentiation of human somatic cells to induced pluripotent stem cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64*.Anokye-Danso F, Trivedi CM, Juhr D, Gupta M, Cui Z, Tian Y, Zhang Y, Yang W, Gruber PJ, Epstein JA, et al. Highly Efficient miRNA-Mediated Reprogramming of Mouse and Human Somatic Cells to Pluripotency. Cell Stem Cell. 2011;8:376–388. doi: 10.1016/j.stem.2011.03.001. The authors dedifferentiate both murine and human somatic cells into induced pluripotent stem cells using the microRNA cluster miR-302/367 alone. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Card DA, Hebbar PB, Li L, Trotter KW, Komatsu Y, Mishina Y, Archer TK. Oct4/Sox2-regulated miR-302 targets cyclin D1 in human embryonic stem cells. Mol Cell Biol. 2008;28:6426–6438. doi: 10.1128/MCB.00359-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cheng LC, Pastrana E, Tavazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci. 2009;12:399–408. doi: 10.1038/nn.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu K, Liu Y, Mo W, Qiu R, Wang X, Wu JY, He R. MiR-124 regulates early neurogenesis in the optic vesicle and forebrain, targeting NeuroD1. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkq904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mavrakis KJ, Wolfe AL, Oricchio E, Palomero T, de Keersmaecker K, McJunkin K, Zuber J, James T, Khan AA, Leslie CS, et al. Genome-wide RNA-mediated interference screen identifies miR-19 targets in Notch-induced T-cell acute lymphoblastic leukaemia. Nat Cell Biol. 2010;12:372–379. doi: 10.1038/ncb2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cottonham CL, Kaneko S, Xu L. miR-21 and miR-31 converge on TIAM1 to regulate migration and invasion of colon carcinoma cells. J Biol Chem. 2010;285:35293–35302. doi: 10.1074/jbc.M110.160069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schramedei K, Morbt N, Pfeifer G, Lauter J, Rosolowski M, Tomm JM, von Bergen M, Horn F, Brocke-Heidrich K. MicroRNA-21 targets tumor suppressor genes ANP32A and SMARCA4. Oncogene. 2011 doi: 10.1038/onc.2011.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zheng J, Xue H, Wang T, Jiang Y, Liu B, Li J, Liu Y, Wang W, Zhang B, Sun M. miR-21 downregulates the tumor suppressor P12 CDK2AP1 and stimulates cell proliferation and invasion. J Cell Biochem. 2011;112:872–880. doi: 10.1002/jcb.22995. [DOI] [PubMed] [Google Scholar]
- 72.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 73.Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 2008;283:14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D, Diehn M, Liu H, Panula SP, Chiao E, et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138:592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hyun S, Lee JH, Jin H, Nam J, Namkoong B, Lee G, Chung J, Kim VN. Conserved MicroRNA miR-8/miR-200 and its target USH/FOG2 control growth by regulating PI3K. Cell. 2009;139:1096–1108. doi: 10.1016/j.cell.2009.11.020. [DOI] [PubMed] [Google Scholar]
- 77.Iliopoulos D, Lindahl-Allen M, Polytarchou C, Hirsch HA, Tsichlis PN, Struhl K. Loss of miR-200 inhibition of Suz12 leads to polycomb-mediated repression required for the formation and maintenance of cancer stem cells. Mol Cell. 2010;39:761–772. doi: 10.1016/j.molcel.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chan YC, Khanna S, Roy S, Sen CK. miR-200b targets Ets-1 and is down-regulated by hypoxia to induce angiogenic response of endothelial cells. J Biol Chem. 2011;286:2047–2056. doi: 10.1074/jbc.M110.158790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Qi J, Yu JY, Shcherbata HR, Mathieu J, Wang AJ, Seal S, Zhou W, Stadler BM, Bourgin D, Wang L, et al. microRNAs regulate human embryonic stem cell division. Cell Cycle. 2009;8:3729–3741. doi: 10.4161/cc.8.22.10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sinkkonen L, Hugenschmidt T, Berninger P, Gaidatzis D, Mohn F, Artus-Revel CG, Zavolan M, Svoboda P, Filipowicz W. MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat Struct Mol Biol. 2008;15:259–267. doi: 10.1038/nsmb.1391. [DOI] [PubMed] [Google Scholar]
- 81.Benetti R, Gonzalo S, Jaco I, Munoz P, Gonzalez S, Schoeftner S, Murchison E, Andl T, Chen T, Klatt P, et al. A mammalian microRNA cluster controls DNA methylation and telomere recombination via Rbl2-dependent regulation of DNA methyltransferases. Nat Struct Mol Biol. 2008;15:998. doi: 10.1038/nsmb0908-998b. [DOI] [PubMed] [Google Scholar]
- 82.Benetti R, Gonzalo S, Jaco I, Munoz P, Gonzalez S, Schoeftner S, Murchison E, Andl T, Chen T, Klatt P, et al. A mammalian microRNA cluster controls DNA methylation and telomere recombination via Rbl2-dependent regulation of DNA methyltransferases. Nat Struct Mol Biol. 2008;15:268–279. doi: 10.1038/nsmb.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lin SL, Chang DC, Lin CH, Ying SY, Leu D, Wu DT. Regulation of somatic cell reprogramming through inducible mir-302 expression. Nucleic Acids Res. 2011;39:1054–1065. doi: 10.1093/nar/gkq850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 85.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 86.Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21:1025–1030. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lin YC, Hsieh LC, Kuo MW, Yu J, Kuo HH, Lo WL, Lin RJ, Yu AL, Li WH. Human TRIM71 and its nematode homologue are targets of let-7 microRNA and its zebrafish orthologue is essential for development. Mol Biol Evol. 2007;24:2525–2534. doi: 10.1093/molbev/msm195. [DOI] [PubMed] [Google Scholar]
- 88.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhao C, Sun G, Li S, Lang MF, Yang S, Li W, Shi Y. MicroRNA let-7b regulates neural stem cell proliferation and differentiation by targeting nuclear receptor TLX signaling. Proc Natl Acad Sci U S A. 2010;107:1876–1881. doi: 10.1073/pnas.0908750107. [DOI] [PMC free article] [PubMed] [Google Scholar]