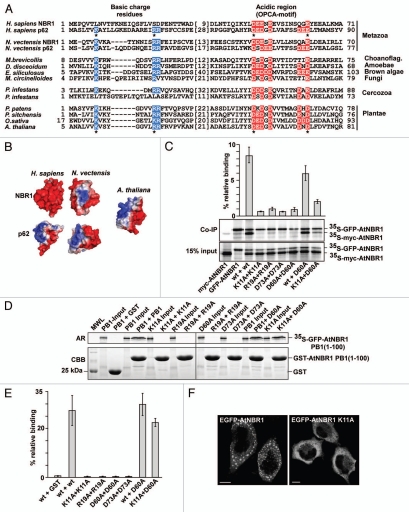
Figure 2.
AtNBR1 polymerizes via the N-terminal PB1-domain. (A) Alignment of PB1 domain sequences from p62- and NBR1 homologs of selected metazoan- and nonmetazoan species. Blue background color denotes basic residues and red background color denotes acidic residues of the charged clusters important for PB1 domain interactions. The OPCA-motif is indicated. Gaps are indicated with dashes and the numbers of amino acid residues not shown are specified in brackets. Residues crucial for the self-interaction of p62,34 and which are mutated in AtNBR1 in the analyses shown in (C–E) are indicated with asterisks. (B) Electrostatic surface potentials of the PB1 domains of NBR1 (PDB:2BKF) and p62 (PDB:2KKC) from H. sapiens and N. vectensis as well as the AtNBR1 PB1 domain. The N. vectensis and AtNBR1 PB1 domains were modeled using Swissmodel. (C) Co-immunoprecipitation experiments using full-length AtNBR1. Myc- and GFP-tagged AtNBR1 (wt or the indicated mutants) were co-translated in vitro in the presence of 35S-methionine and precipitated using an anti-GFP antibody. Immunoprecipitated and co-precipitated proteins as well as in vitro translated proteins corresponding to 15% of the input were resolved by SDS-PAGE and detected by autoradiography. The upper band corresponds to 35SGFP-AtNBR1, the lower to 35Smyc-AtNBR1. Quantifications of the interaction data are shown above the gel parts. Band intensity was measured using ImageJ (Fuji) and Y-axis values were calculated employing the following formula; (IP(myc/GFP)/input(myc/GFP)) × 100. (D) GST pulldown assays using in vitro translated 35S-labeled GFP-AtNBR1 PB1 (amino acids 1–100) (with the indicated point mutations) and GST or GST-PB1 (with indicated point mutations) constructs. Precipitated proteins were detected by autoradiography. (E) Quantitative representation of the interaction data shown in (D). Y-axis values are set to percent total binding protein; (pulldown/input) × 100. (F) AtNBR1 forms cytosolic aggregates when overexpressed with an N-terminal GFP-tag in HeLa cells, while the K11A point-mutant of AtNBR1 loses the ability to form aggregates. Results in (C and E) are mean values of three independent experiments with standard deviations indicated as bars. Bars represent 10 µm.