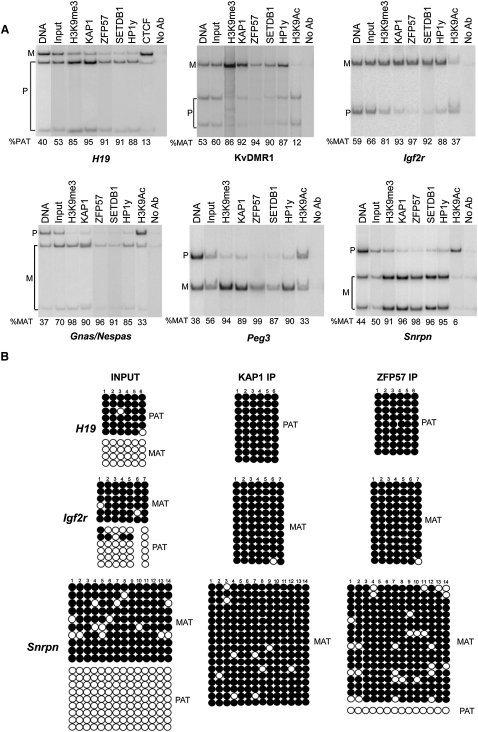
Figure 3.
KAP1, ZFP57, SETDB1, and HP1 Are Selectively Recruited to the Methylated Alleles of Imprinted Control Regions
(A) RFLP analysis of KAP1-, ZFP57-, SETDB1- and HP1-bound material. Following ChIP with indicated antibodies recognizing endogenous proteins and ICR-specific PCR amplification, products were digested with restriction enzymes that cut only one parental allele to measure the ratio of maternal (%MAT) or paternal (%PAT) allele relative to total. H3K9me3 (which is known to bind to the methylated ICRs allele) and H3K9Ac (which is known to bind to the unmethylated ICRs allele) were analyzed as control. No Ab, no specific primary antibody.
(B) Bisulfite sequencing of the KAP1- and ZFP57- bound H19, Igf2r and Snrpn ICRs. Methylated and unmethylated CpG dinuclotides are represented as open and filled circles, respectively. The input (pre-ChIP) DNA shows a 50/50 ratio of methylated and unmethylated ICR, although the KAP1- and ZFP57-immunoprecipitated (IP) material is totally or prevalently methylated. Each line corresponds to a single template DNA molecule cloned; each circle corresponds to a CpG dinucleotide. Filled circles designate the methylated cytosines; opened circles unmethylated cytosines. Differences are statistically significant (P-values < 0.05).