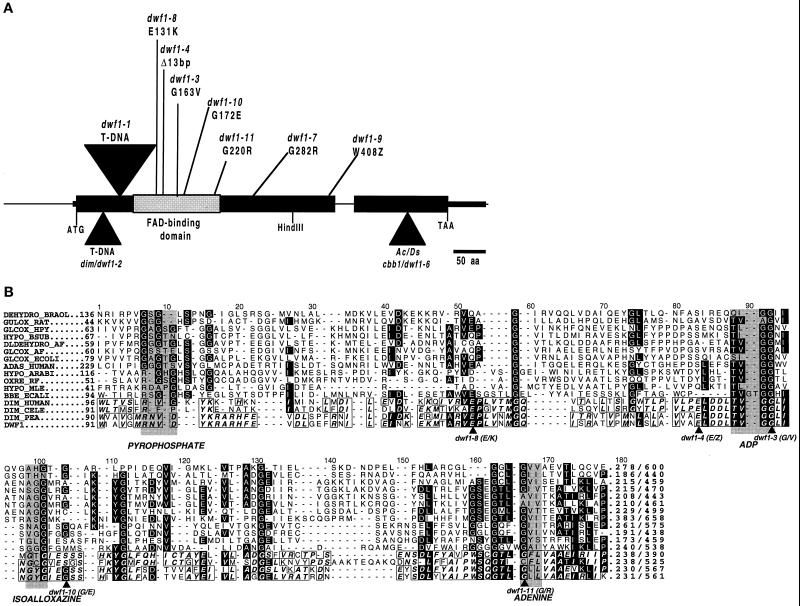
Figure 5.
Schematic representation of Arabidopsis DWF1 locus with characterized mutations (A) and aligned amino acid sequences of proposed FAD-binding domains from various oxidoreductases (B). Elucidation of DWF1 organization resulted from comparison of cDNA with genomic DNA. Sites of the FAD-binding domain (Mushegian and Koonin, 1996; Fraaije et al., 1998) and mutations identified from dwf1 alleles, including dim (dwf1-2; Takahashi et al., 1995) and cbb1 (dwf1-6; Kauschmann et al., 1996), are indicated. Sequences corresponding to the region of the FAD-binding domain depicted in A are aligned to show conserved residues and relative positions of mutations. Accession numbers for the following are given in parentheses: DEHYD_BRAOL (2760543), GULOX_RAT (625202), GLCOX_HPY (2313619), HYPO_BSUB (1770026), DLDEHYD_AF (2650235), GLCOX_AF (2649802), GLCOX_ECOLI (1707917), ADAS_HUMAN (2498106), HYPO_ARABI (2618686), OXRE_RF (1169648), HYPO_MLE (3150105), BBE_ECALI (400972), DIM_HUMAN (3182980), DIM_CELE (3182979), DIM_PEA (3182981), DWF1 (U12400). Similar sequences were identified using gapped BLAST (Altschul et al., 1997), followed by alignment using PileUp software (GCG). Box shading was carried out using the ALSCRIPT package developed by Barton (1993).