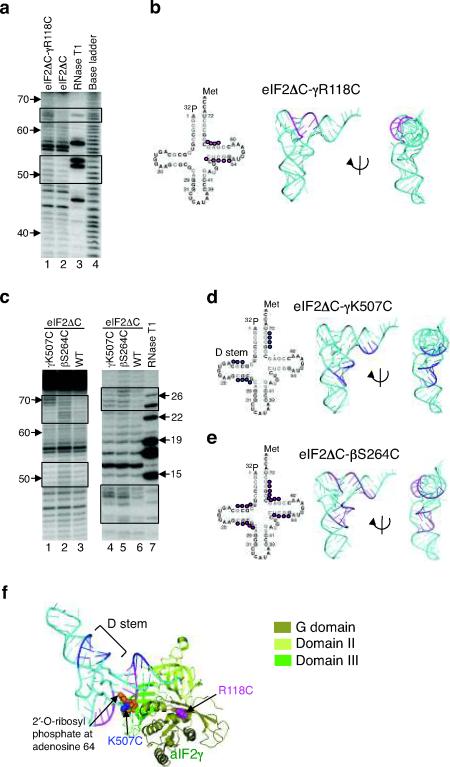
Figure 3.
Directed hydroxyl radical cleavage of Met-[32P]tRNAiMet by Fe(II)-BABE-derivatized eIF2 in 48S complexes.
(a) eIF2ΔC-γR118C-Fe(II)-BABE cleavages. Hydroxyl radical cleavage products from 48S complexes were resolved on 10% (w/v) denatured polyacrylamide gels, and cleavage sites on [32P]tRNAiMet were determined by comparison with samples containing eIF2ΔC (WT) (lane 2). Sites of enhanced cleavage with eIF2ΔC-R118C are boxed. The tRNA ladders were prepared by digesting Met-[32P]tRNAiMet with RNase T1 (cleaves 3′ of G residue, lane 3) or by base cleavage (lane 4). The tRNA residue numbers are shown at the left.
(b) Sites (colored magenta) of eIF2ΔC-γR118C-Fe(II)-BABE directed hydroxyl radical cleavage on Met-[32P]tRNAiMet are shown on the secondary (left) and three-dimensional (pdb code: 1YFG19, middle and right) structures of tRNAiMet.
(c) Met-[32P]tRNAiMet cleavages in 48S complexes assembled with eIF2ΔC-γK507C-Fe(II)-BABE and eIF2ΔC-βS264C-Fe(II)-BABE, as in panel a. Cleavage products were resolved on 10% (left) or 20% (right) (w/v) denatured polyacrylamide gels.
(d,e) Summary of Met-[32P]tRNAiMet cleavages in panel c, depicted as in panel b.
(f) aIF2γ-GDPNP-tRNAiMet TC model showing the sites of tRNAiMet (cyan) cleavages by hydroxyl radicals generated at residues K507 (blue) and R118 (magenta). The model was generated by docking the structure of yeast tRNAiMet (pdb code: 1YFG19) on the aIF2γ TC structure from Fig. 1b. The RIT1 catalyzed 2′-O-ribosyl phosphate modification at residue 64 of tRNAiMet is shown as orange spheres.