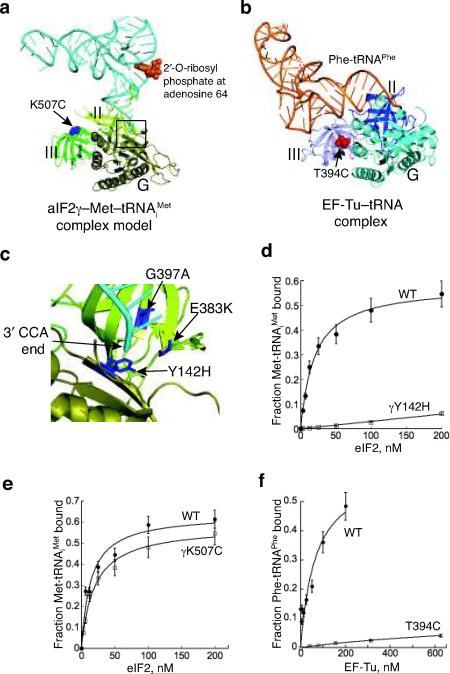
Figure 5.
eIF2γ binding site for 3′ end of Met-tRNAiMet.
(a) aIF2γ–Met-tRNAiMet complex model, generated as described in Figs. 4c–d. The RIT1 catalyzed 2′-O-ribosyl phosphate modification at residue 64 of tRNAiMet is shown in orange.
(b) Ribbons representation of EF-Tu–GDPNP–Phe-tRNAPhe complex (pdb code: 1TTT18). The site of the T394C mutation, which is analogous to the eIF2γ-K507C mutation, is shown as red spheres.
(c) Magnified view of proposed eIF2γ binding site for the 3′ end of Met-tRNAiMet (box in Fig. 5a). Locations of yeast eIF2γ mutations that impair eIF2 function in vivo are labeled and shown as blue sticks: Y142H (corresponds to S. sol. aIF2γ-Tyr51), E383K (Gly282), and G397S (Ala296).
(d,e) Purified WT, γY142H (D), or γK507C (E) mutant forms of eIF2 were assayed for eIF2 TC formation by filter binding assay. Fractions of [35S]Met-tRNAiMet bound to eIF2 were plotted as a function of eIF2 concentration; points and s.d. are averages of at least three independent experiments.
(f) Purified WT or T439C mutant forms of EF-Tu (T. therm) were assayed for TC formation by filter binding assay. Fractions of [14C]Phe-tRNAPhe bound to EF-Tu were plotted as a function of EF-Tu concentration; points and s.d. are averages of at least three independent experiments.