Figure 1.
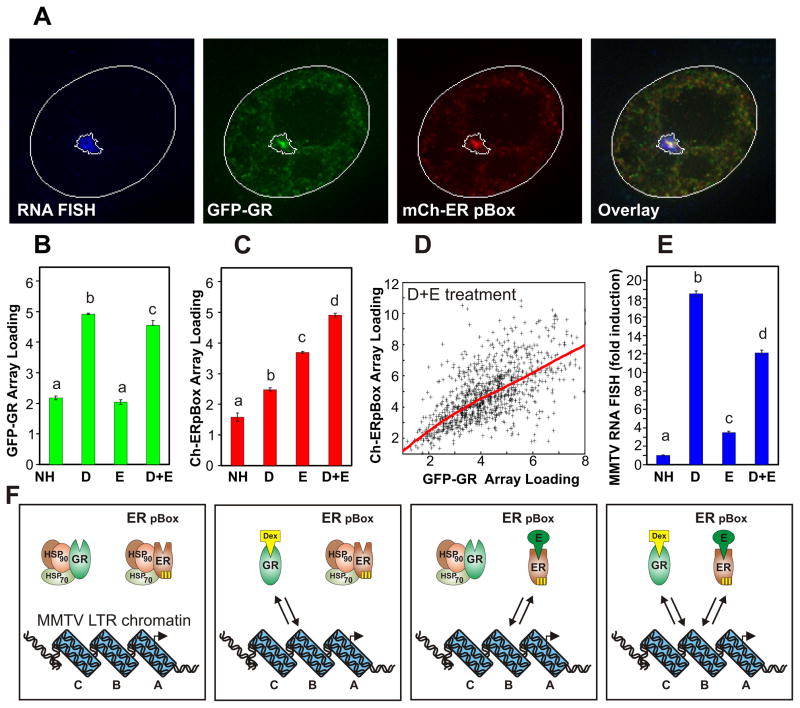
Single cell level analysis of competitive steroid receptor interactions with chromatin. Treatment with Dex or E2 allows DNA-binding of GR or ER pBox, respectively. (A) 6644 cell line cultures were treated for 30 minutes with both Dex and E2, then processed for RNA FISH to detect transcription from the MMTV array. The micrographs display a single nucleus with each fluorescence channel shown separately and merged in the overlay. Computationally defined ROIs for the nucleus and MMTV array FISH signal are outlined in white. (B–D) Based on automatic detection of the MMTV array FISH signal in large numbers of individual cells (n = 500 to 1000 per condition), steady-state receptor interactions with the MMTV array are measured following 30m hormone treatment. (D) The points of the scatter plot show the relative amount of GR and ER pBox interaction with the MMTV array for individual cells within the Dex+E2 treated population, and the red line represents best fit relationship between these parameters. (E) The image analysis algorithm also simultaneously measures MMTV RNA FISH signal. Bars represent the mean of cellular measurements, error bars denote SEM, and markers “a,b,c” indicate homogenous statistical subsets of conditions. Each subset is considered to be statistically different because it consists of measurements with HSD p-values < 0.05 versus the values in another defined subset. (F) Diagrams illustrate the dynamic interaction of GR protein and/or ER pBox protein with the chromatinized GRE located in the B-nucleosome region of the MMTV-LTR promoter. See also Supplemental Figure 1.