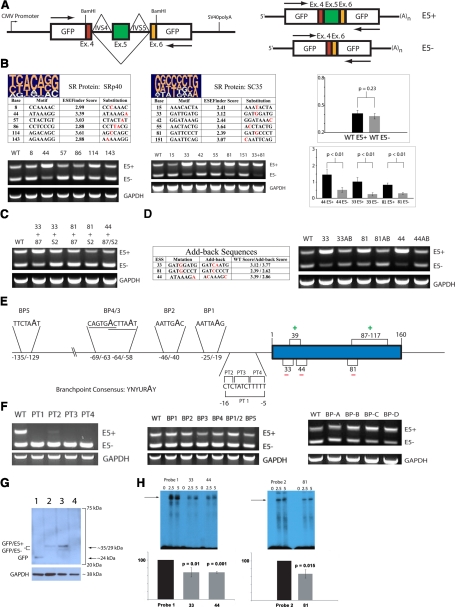
Figure 2.
Spliceosomal definition of TF exon 5: SR protein-dependent mechanisms predominate. (A) Schematic of the reporter system pGL-hTF. Arrows indicate positions of primers used in conventional RT-PCR. Exon 5-containing amplicon (E5+), 432 bp; exon 5-lacking amplicon (E5-), 272 bp. (B) SRp40 promotes TF exon 5 exclusion via the ESS at position 44 and SC35 via ESS at positions 33 and 81. At the top of each table are pictograms of the degenerate motifs for SRp40 and SC35 [9]; mutagenized positions in each putative site are shown in red. Representative RT-PCR (n≥3) of the resultant expression patterns is below each table. In the graphs, the change in the degree of exon 5 definition as a result of ESS weakening results in the change of the splicing pattern. At least three independent transfection assays were normalized to GAPDH, quantified using ImageJ software v.1.42, and averaged; error bars are sd. WT, Wild-type. (C) Representative RT-PCR (n≥4), the expression pattern of the combinatorial mutants featuring weakened ASF/SF2 ESE and weakened SRp40/SC35 ESS. (D) Representative RT-PCR (n≥4), expression pattern of the “add-back” mutants whose sequences and scores are shown in the table, featuring nonendogenous SRp40 and SC35 ESS. (E) The diagram depicting the opposing roles of SR proteins in TF exon 5 definition and the potential structural elements in TF intron 4. (F, left panel) Representative RT-PCR (n≥4), the expression pattern of the pGL-hTF mutants featuring purine substitutions in the PT of intron 4. (Middle panel) Representative RT-PCR (n≥4), the expression pattern of the pGL-hTF mutants featuring adenine-to-guanine substitutions in the potential BP motifs of intron 4. (Right panel) Representative RT-PCR (n≥4), pGL-hTF mutants featuring multiple adenine-to-guanine substitutions clustered within the last 100 bases of intron 4. (G) Representative Western blot (n≥3), THP-1 was transiently transfected for 60 h with pGL (Lane 1), pGL-hTF wild-type (Lane 2), pGL-hTF harboring weakened SC35 sites 33 and 81 (Lane 3), and pGL-hTF harboring an expensively weakened PT of exon 5 (PT1, Lane 4), lysed and probed with anti-GFP mAb (Roche Life Sciences, Indianapolis, IN, USA). The blot was stripped and reprobed with polyclonal anti-GAPDH antibodies as described [10]. (H, upper panels) Representative radiograms (n=3) of RNA mobility shift assays using wild-type and mutagenized probes featuring the ESS 33, 44, and 81. Numbers on top indicate the amounts of nuclear protein (in μg). (Lower panels) The results of three independent assays were quantified as described in Materials in Methods and averaged. (In both graphs, the intensity of the major RNA-protein complexes produced by wild-type probes was treated as 100%; error bars are sd.)