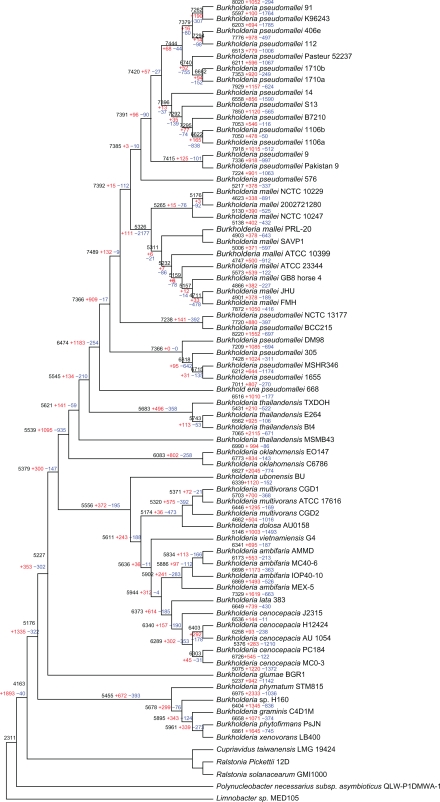
Figure 1.
Phylogenetic tree of Burkholderiaceae based on concatenated amino acid alignments of one homolog gene and rooted by using proteins from other Burkholderiaceae species as the outgroup. All the evolutionary nodes in this tree were supported by bootstrap values higher than 90%. Reconstruction of gene content evolution based on BuCOGs in Burkholderia is shown. For each species and each internal node of a tree, the inferred number of BuCOGs present (numbers in black), and the numbers of BuCOGs lost (numbers in blue) or gained (numbers in red) along the branch leading to a given node (species) are indicated.