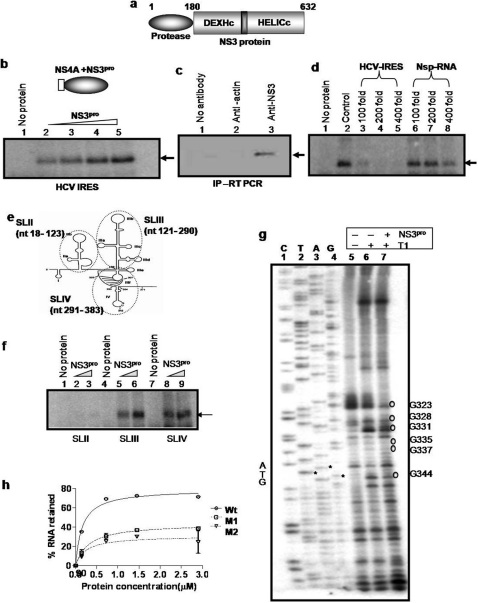
Figure 1. NS3pro specifically binds to HCV IRES RNA.
Panel a: Schematic representation of the domain organization of HCV NS3 protein. Panel b: UV-crosslinking. [α32P]UTP labeled HCV IRES RNA was UV cross linked with increasing concentrations (0.1, 0.2, 0.4 and 0.8 µM) of NS3pro (lane 2 –5). Lane 1 shows only probe control. Schematic representation of the NS3pro protein is shown above the panel. Panel c: Immuno-pulldown assay and RT-PCR. NS3-RNA complex was immunoprecipitated from HCV replicon bearing cells using anti-NS3 antibody. RNA was isolated and RT-PCR was performed for HCV IRES (lane 3). No antibody (lane 1) and anti-actin antibodies (lane 2) were used as negative controls for the pull down. Panel d: Competition UV-crosslinking. [α32P]UTP labeled HCV IRES RNA was UV cross linked with NS3pro in the absence (lane 2) and presence of molar excess of unlabeled HCV IRES RNA (lanes 3–5) or a nonspecific RNA (lanes 6–8). Lane 1 shows only probe control. Panel e: Schematic representation of HCV IRES RNA showing the stem and loop (SL) regions 20. The domains that are used as SLII, III and IV in the study are encircled and indicated. Panel f: UV-crosslinking. [α32P]UTP labeled HCV SLII (lane 2–3); SLIII (lane 5–6) and SLIV (lane 8–9) RNAs were UV cross linked to increasing concentrations (0.2 and 0.4µM) of NS3pro. Lanes 1, 4 and 7 represents only probe controls for SLII, SLIII and SLIV respectively. Panel g: RNase T1 Foot-printing assay. Binding reactions of in vitro transcribed HCV IRES RNA were carried out in absence (lane 6) or presence (lane 7) of NS3 protease. The RNA was then digested with RNase T1. RNA was reverse-transcribed with an end labeled primer. The cDNA was resolved in along with a reference sequencing reaction (lanes 1–4). Lane 5 represents the no T1 control. Panel h: Filter-binding assay. [α32P]UTP labeled HCV IRES RNA or mutant IRES RNA (M1 and M2) was bound to increasing concentrations of NS3pro.