Fig. 4.
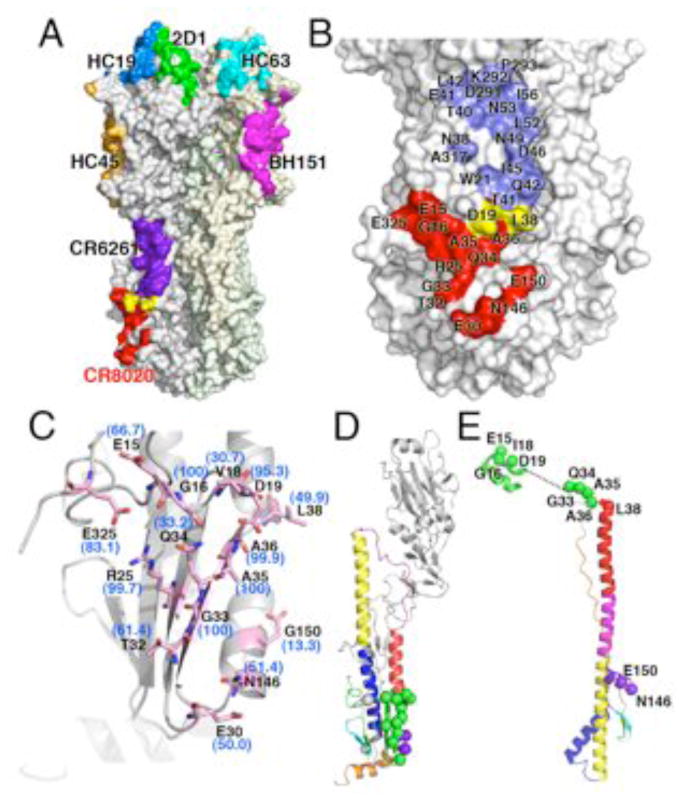
CR8020 binds a unique, highly conserved site in the stem. (A) Comparison of CR8020 binding site to the epitopes of all other structurally characterized antibodies. All antibody footprints are mapped onto the surface of HK68 HA, although they are derived from different structures (and subtypes): Red, CR8020 (PDB code: XXXX); Purple, CR6261 (PDB codes 3GBM and 3GBN); green, 2D1 (PDB code: 1LFZ); blue, HC19 (PDB code: 2VIR); orange, HC45 (PDB code: 1QFU); cyan, HC63 (PDB code: 1KEN); pink, BH151 (PDB code: 1EO8). Note that mAb F10 and CR6261 bind the same epitope in the stem, but only CR6261 is illustrated here for clarity. (B) Comparison of the broadly neutralizing CR8020 and CR6261 epitopes, which constitute discrete surfaces on on group 2 and group1 HAs, respectively. Coloring is similar to (A), with CR8020 epitope in red, CR6261 epitope in blue, and shared epitope residues in yellow. (C) Conservation of CR8020 epitope across group 2 HAs. Residues comprising the epitope are shown as sticks (carbon in light pink, oxygen in red and nitrogen in blue). Percent identity with the group 2 consensus sequence is indicated alongside each residue. Note that the residue label reflects the most common residue across group 2 HAs, which is not always identical to the residue at that position in the HK68 crystal structure. View is similar to that of Fig. 4B, looking from the Fab (not shown) towards the epitope. (D) Location of CR8020 epitope residues in the pre-fusion and (E) post-fusion state, where they reside on critical regions of the fusion peptide and helix-capping region of HA2. For D and E, CR8020 epitope residues are indicated as spheres, with green mapping to the fusion peptide and N-cap region. Additional contact residues are purple (HA2) and gray (HA1, not present in available post-fusion structures). Residue positions were mapped onto the structures from PDB codes 1QU1 and 2KXA.
