Abstract
Association of herpesvirus DNA with histones has important implications for lytic and latent infection; thus herpesviruses arbitrate interactions with histones to productively infect host cells. While regulation of alpha and betaherpesvirus chromatin during lytic infection has been actively investigated, very little is known about interaction of gammaherpesvirus DNA with histones upon de novo lytic infection. Murine gammaherpesvirus-68 (MHV68) is a rodent pathogen that offers a tractable system to study gammaherpesvirus lytic infection in primary cells. In this study we report that MHV68 promoter and orilyt sequences underwent dynamic association with histone H3 during de novo lytic infection of primary macrophages and fibroblasts. Similar to HSV-1, the degree of MHV68 DNA association with histone H3 was dependent on the multiplicity of infection and was further regulated by viral DNA synthesis. Our work sets a precedent for future studies of gammaherpesvirus chromatin during de novo lytic infection.
Keywords: gammaherpesvirus, chromatin, core histone
Introduction
Eukaryotic DNA is organized as chromatin, primarily via interaction of DNA with nucleosomes. The nucleosome is composed of core histones H2A, H2B, H3, and H4, which wrap approximately 147 bp of DNA. Chromatin structure is dynamic and plays an important role in DNA replication and transcription.
Herpesviruses are large, double-stranded DNA viruses that replicate in the nucleus of host cells and rely on host transcriptional machinery for viral gene expression. The association of herpesvirus DNA with histones has profound effects on viral gene expression and control of replication and latency [reviewed in (Knipe and Cliffe, 2008;Lieberman, 2008;Paulus et al., 2010)]. Unlike small DNA viruses that package histone-associated DNA (Favre et al., 1977;McMillen and Consigli, 1974), herpesvirus virion DNA is histone-free and instead associates with polyamines (Gibson and Roizman, 1971;Cohen et al., 1980;Oh and Fraser, 2008;Pignatti and Cassai, 1980). Upon infection, Herpes Simplex Virus-1 (HSV-1) and Human Cytomegalovirus (HCMV) DNA rapidly associates with cellular histones (Kent et al., 2004;Oh and Fraser, 2008;Nitzsche et al., 2008;Cuevas-Bennett and Shenk, 2008;Cliffe and Knipe, 2008). The precise mechanism by which histones are loaded on herpesvirus DNA has not been elucidated; however, both viral proteins and cellular histone chaperones are likely to be involved (Ambagala et al., 2009;Cliffe and Knipe, 2008;Placek et al., 2009;Kutluay and Triezenberg, 2009;Peng et al., 2010). Furthermore, lower multiplicity of infection (MOI) results in higher levels of histones associated with HSV-1 DNA early in infection (Cliffe and Knipe, 2008). While core histones remain associated with the HCMV genome throughout lytic replication (Cuevas-Bennett and Shenk, 2008;Nitzsche et al., 2008), association of core histones with HSV-1 DNA decreases during viral DNA synthesis; this decrease is most pronounced with respect to newly synthesized viral DNA (Cliffe and Knipe, 2008;Oh and Fraser, 2008). The mechanism by which histones are removed and/or prevented from association with HSV-1 DNA is not well established.
While a wealth of reports describes dynamic histone-viral DNA interactions during alpha- and betaherpesvirus lytic infection, very little is known about the association of gammaherpesvirus DNA with histones in the context of de novo lytic infection (Gunther and Grundhoff, 2010). Understanding gammaherpesvirus infection of naïve cells is highly relevant as de novo infection is important during acute replication and for the maintenance of the pool of infected cells in chronically infected host in vivo (Liang et al., 2009;Hoshino et al., 2009). Unfortunately, exquisite species specificity of human gammaherpesviruses Epstein-Barr virus (EBV) and Kaposi’s sarcoma associated herpesvirus (KSHV) limits studies of de novo lytic infection in primary cells. MHV68 is a rodent pathogen that is genetically and biologically related to human EBV and KSHV (Efstathiou et al., 1990;Virgin et al., 1997). MHV68 represents a robust experimental system to explore parameters of de novo lytic infection of primary macrophages, a physiologically-relevant cell type that supports both lytic and latent MHV68 infection (Tarakanova et al., 2007;Weck et al., 1999). In this report we show that MHV68 promoter and orilyt sequences underwent dynamic association with core histone H3 during lytic replication in primary macrophages and mouse embryonic fibroblasts, with the latter representing a cell type where MHV68 lifestyle is limited to lytic replication. The dynamics of MHV68 DNA-histone H3 association were closer to that observed during HSV-1, and not HCMV, infection, including MOI-dependent effects on histone association. Furthermore, viral DNA synthesis was not required for the partial removal of histone H3 from MHV68 DNA sequences of interest, suggesting that dissociation of histone H3 from viral DNA was programmed during an early stage of infection. Our studies set a precedent for future investigations of viral and host factors involved in regulation of gammaherpesvirus chromatin during de novo lytic infection.
Results and Discussion
MHV68 DNA sequences undergo dynamic association with histone H3 during de novo lytic infection of primary bone marrow macrophages
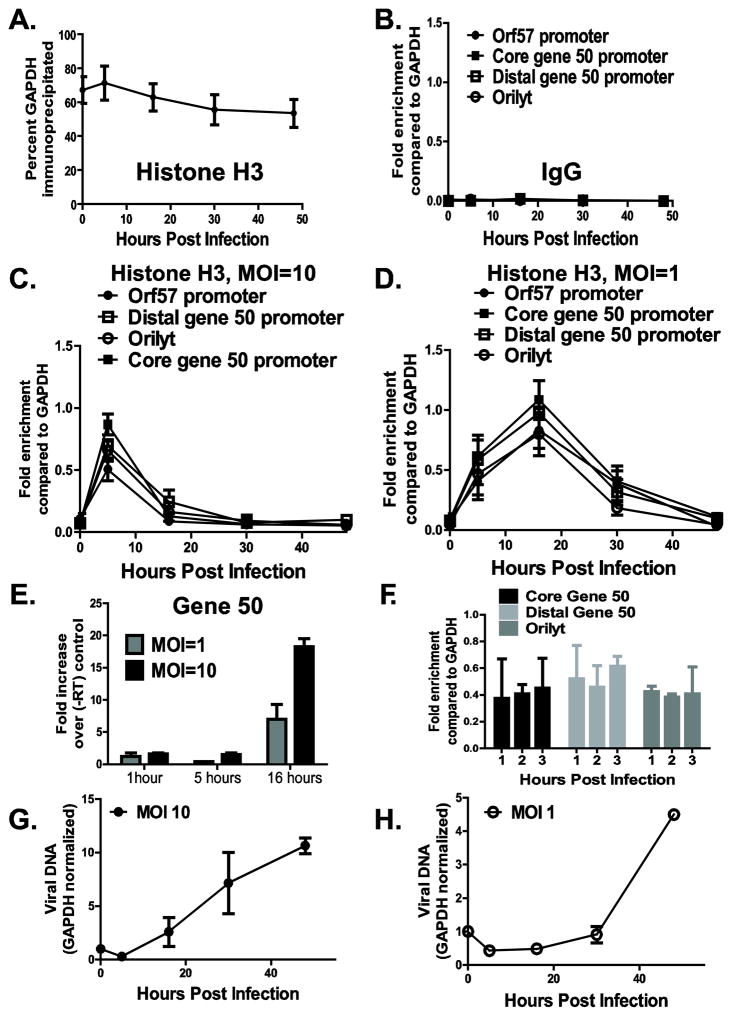
To determine whether MHV68 DNA was associated with core histones during lytic infection, primary bone marrow derived macrophages were infected with wild type MHV68, chromatin harvested throughout a single cycle of replication (48 hours) and subjected to chromatin immunoprecipitation (ChIP) using anti-histone H3 or IgG. DNA levels of several MHV68 promoters and the right origin of lytic replication (orilyt) (Deng et al., 2004;Gray et al., 2008;Liu et al., 2000) were measured by quantitative real time PCR in input and immunoprecipitated samples, and normalized to corresponding GAPDH levels. On average, approximately 60% of GAPDH sequences were immunoprecipitated with anti-histone H3 antibody (Fig. 1A). Association of GAPDH with histone H3 remained constant throughout a single cycle of replication (Fig. 1A), validating the use of GAPDH for normalization. Furthermore, immunoprecipitation with an negative control antibody (IgG) failed to enrich for viral or host genomic sequences as compared to sequences immunoprecipitated from the same chromatin samples with specific anti-histone H3 antibody (Fig. 1B), suggesting that DNA sequences probed do not nonspecifically associate with agarose beads during immunoprecipitation.
Figure 1. MHV68 DNA sequences undergo dynamic association with histone H3 during de novo lytic infection of primary bone marrow macrophages.
Primary bone marrow macrophages were infected at an indicated MOI with wild type MHV68, and chromatin was harvested throughout a single cycle of replication and subjected to ChIP using anti-histone H3 or a negative control antibody (IgG), as indicated (A–D, F). Efficiency of immunoprecipitation of indicated MHV68 DNA sequences was normalized to the efficiency of immunoprecipitation of GAPDH (A) using the ΔΔCt method and is presented as relative enrichment over GAPDH (B–D, F). E. Total RNA was harvested at indicated times post infection and converted to cDNA in the presence or absence of reverse transcriptase (RT). Abundance of RTA sequences was measured by real time PCR and is presented as fold difference between the corresponding plus RT and minus RT samples. F. Primary macrophages were infected at an MOI of 1, chromatin harvested at indicated times, and subjected to ChIP using anti-histone H3 antibody. G, H. Viral DNA was measured in input samples by real time PCR at indicated time points post infection and normalized to corresponding GAPDH levels. For each panel, data were pooled from 2–5 independent experiments, with error bars representing one standard error of the mean.
Very little viral DNA was associated with histone H3 immediately post absorption (0h, Fig. 1C, D), consistent with previous reports of non-histone based herpesvirus DNA packaging in the virion (Cohen et al., 1980;Oh and Fraser, 2008;Pignatti and Cassai, 1980). The enrichment of MHV68 sequences immunoprecipitated with the anti-histone H3 antibody as compared to the negative control IgG at 0h post infection was likely due to incomplete digestion of unpackaged viral DNA during MHV68 stock preparation. At 5h post infection, all examined MHV68 sequences were associated with histone H3, regardless of the MOI (Fig. 1C, D). The extent of histone H3 association with MHV68 sequences was lower than that observed for H3 association with cellular sequences (25–50% of viral sequences were immunoprecipitated with anti-histone H3 as compared to 70% of cellular GAPDH immunoprecipitated at this time point, Fig. 1A, C, D). This reduced association may result from a heterogenous population of chromatinized genomes or irregular chromatinization of the examined viral promoters. The levels of immediate early gene 50 transcripts were below the threshold of detection at 1 and 5 hours post infection (Fig. 1E), suggesting that the rapid chromatization of MHV68 sequences occurred prior to active viral gene transcription and may have been facilitated by viral tegument and/or cellular proteins.
Having detected significant association of all examined MHV68 sequences with histone H3 at 5h post infection, we wanted to examine additional time points between 0 and 5h post infection. Surprisingly, a significant amount of examined MHV68 DNA sequences associated with histone H3 as early as 1h post infection (Fig. 1F, compare fold enrichment of 0.4–0.6 at 1h post infection to fold enrichment of less than 0.1 at 0h post infection, Fig. 1D), suggesting that delivery of viral DNA to the nucleus and its association with histones initiates within the first hour following viral entry.
Between 5 and 16h post infection, a decrease in association of histone H3 with MHV68 sequences was observed under high MOI conditions (Fig. 1C). This decrease happened concurrent with the initiation of MHV68 DNA synthesis (Fig. 1G). In contrast, histone H3 levels bound to MHV68 DNA further increased between 5 and 16h post infection under low MOI conditions and prior to viral DNA synthesis (Fig. 1D) to match the levels of histone H3 associated with the GAPDH gene (~60% of viral or cellular GAPDH sequences immunoprecipitated with anti-histone H3, Fig. 1A, D). The levels of histone H3 bound to the MHV68 promoters and orilyt decreased by 30h post infection under conditions of low MOI (Fig. 1D). Consistent with the observation for the high MOI, this decrease was coincident with the initiation of viral DNA synthesis (Fig. 1D, H). Very low levels of histone H3 associated with MHV68 DNA sequences at 48h post infection, regardless of the MOI conditions (Fig. 1C, D). Thus, MHV68 promoter and orilyt sequences displayed dynamic association with histone H3 in lytically-infected macrophages.
Histone H3 associates early and dynamically with viral DNA sequences during de novo lytic MHV68 infection in primary murine embryonic fibroblasts
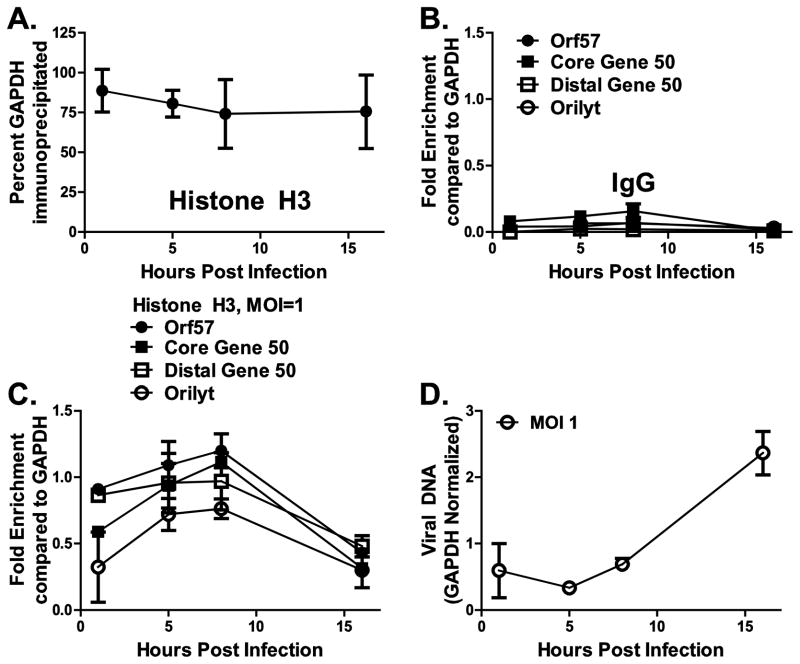
To determine if the dynamics of histone H3 association with viral sequences was unique to infected macrophages, we infected primary murine embryonic fibroblasts (MEFs) at an MOI of 1 and performed chromatin immunoprecipitation with anti-histone H3 and IgG, as described above for primary macrophages. As in macrophages, immunoprecipitation of cellular GAPDH sequences with anti-histone H3 antibodies remained consistent throughout infection (on average, 75% of GAPDH sequences were immunoprecipitated with anti-histone H3, Fig. 2A). Similar to macrophage studies, IgG immunoprecipitated low levels of viral DNA sequences as compared to immunoprecipitation using anti-histone H3 antibody (Fig. 2B and C).
Figure 2. Histone H3 associates early and dynamically with viral DNA sequences in de novo lytic MHV68 infection of primary murine embryonic fibroblasts.
Primary murine embryonic fibroblasts (MEFs) were infected at an MOI of 1 with wild type MHV68, chromatin was harvested at the specified time points during a single round of viral replication, and subjected to ChIP using anti-histone H3 or a negative control antibody (IgG) as indicated (A–C). Efficiency of immunoprecipitation of indicated MHV68 DNA sequences was normalized to the efficiency of immunoprecipitation of GAPDH (A) using the ΔΔCt method and is presented as relative enrichment over GAPDH (B, C). D. Viral DNA was measured in input samples by real time PCR at indicated time points post infection and normalized to corresponding GAPDH levels. For each panel, data were pooled from 2–5 independent experiments, with error bars representing one standard error of the mean.
As early as 1 hpi, orilyt and viral promoter sequences were found to associate with histone H3 to between 40 and 80% of histone H3 level found associated with cellular GAPDH sequences (Fig. 2C). This association increased at 5 and 8 hpi. By 16 hpi, enrichment had decreased to approximately 50% that of cellular GAPDH. The differences in kinetics of viral DNA association with histone H3 in MEFs versus macrophages are consistent with faster MHV68 replication in MEFs: a decrease in histone H3 association occurred concomitantly with viral DNA replication in MEFs (Fig. 2D). Importantly, we observe that in both primary cell types, MHV68 DNA sequences undergo dynamic associations with core histone H3 and that these sequences become less enriched as viral genome replication commences.
Removal of histone H3 from template MHV68 DNA occurs in the absence of viral DNA synthesis
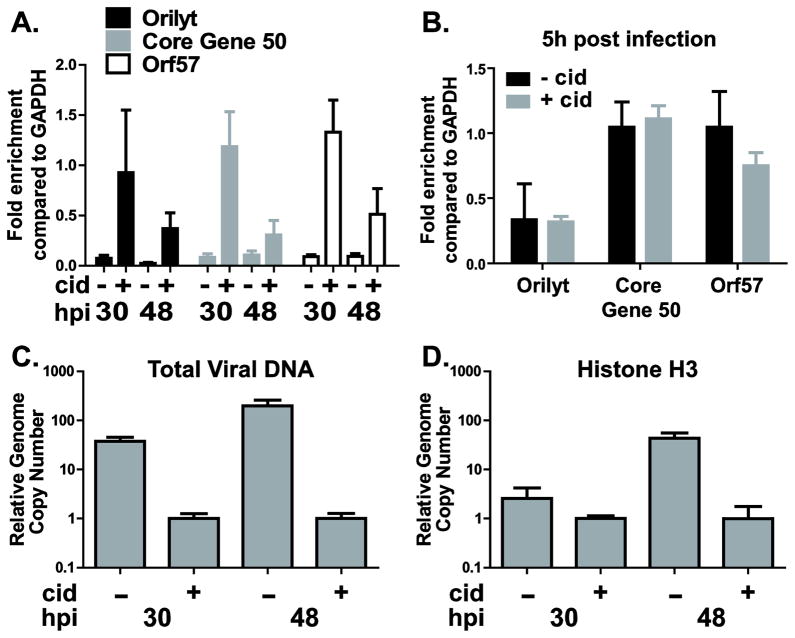
The decrease in histone H3 association with MHV68 sequences observed concurrent with the initiation of viral DNA synthesis could result from an increase in histone-depleted newly synthesized viral DNA (Oh and Fraser, 2008). Alternatively, histone H3 levels could be reduced on both template and newly synthesized viral DNA concurrent with DNA replication, as shown for HSV-1 (Cliffe and Knipe, 2008). To determine the dynamics of histone H3 association with the template MHV68 DNA, macrophages were infected at an MOI of 10 and, immediately after adsorption of virus, treated with 1 μg/ml cidofovir to inhibit viral DNA synthesis (Neyts and De, 1998). Chromatin was collected at several times post infection and subjected to ChIP using anti-histone H3 or a negative control antibody (IgG). As expected, low levels of histone H3 associated with MHV68 DNA in control cells at 30 and 48h post infection (approximately 5–10% of histone H3 levels associated with GAPDH, Fig. 3A). In contrast, high levels of histone H3 remained bound to the template MHV68 DNA in cidofovir-treated cells at 30h post infection, suggesting that an overall decrease in histone H3 association with MHV68 DNA observed in untreated controls at this time point is likely due to decreased association of histone H3 with newly synthesized viral DNA. Importantly, between 30 and 48h post infection a decrease in the levels of histone H3 bound to the examined MHV68 DNA sequences was observed (Fig. 3A). In contrast, cidofovir had no significant effect on the association of histone H3 with several MHV68 DNA sequences at 5h post infection (Fig. 3B). As expected, cidofovir treatment was effective in blocking MHV68 DNA replication throughout the infection (Fig. 3C). Thus, the partial decrease in the levels of histone H3 bound to the MHV68 template DNA between 30 and 48h post infection indicated that H3 clearance from template MHV68 DNA was not exclusively dependent on viral DNA synthesis and late gene expression.
Figure 3. Viral DNA synthesis is not required for partial removal of histone H3 from template MHV68 DNA and does not eliminate association of histone H3 with newly synthesized MHV68 DNA.
Primary bone marrow macrophages were infected at MOI 10 with wild type MHV68 and treated with PBS or 1 g/ml of cidofovir for the duration of the experiment. Chromatin was harvested at 30 and 48h post infection and subjected to ChIP using anti-H3 (A, B). Efficiency of immunoprecipitation of indicated MHV68 DNA sequences was normalized to the efficiency of immunoprecipitation of GAPDH using ΔΔCt method. C. Viral DNA was measured in input samples by real time PCR at indicated time points post infection and normalized to corresponding GAPDH, with relative levels observed at 30h post infection in cidofovir treated cells set to 1. D. Relative number of MHV68 sequences associated with histone H3 under indicated conditions. This number was determined based on efficiency of immunoprecipitation with histone H3 antibody and relative viral DNA levels (calculated in C). For each panel, data were pooled from 2–3 independent experiments and error bars presented as one standard error of the mean.
We have found that viral DNA synthesis coincided with an overall decrease in the levels of histone H3 associated with MHV68 sequences. To determine whether this decrease was due to the lack of histone H3 association with the newly synthesized DNA, we compared relative genome copy numbers of total and histone H3-associated MHV68 DNA under various conditions. At 48h post infection, a 200-fold difference in the relative viral DNA levels was observed between cidofovir treated and control cells (Fig. 3C). However, only a 40-fold difference in the relative number of viral genomes associated with histone H3 was observed under the same conditions (Fig. 3D, 48h post infection, cidofovir treated vs. control groups). These results indicate that although the proportion of viral DNA associated with histone H3 decreased with viral DNA replication (Fig. 3A), the total number of viral genomes associated with histone H3 increased as viral DNA synthesis progressed. Similar differences existed at 30h post infection (Fig. 3C, D). Thus histone H3 associated with both template and newly synthesized MHV68 DNA sequences in primary macrophages, and the relative number of MHV68 sequences associated with histone H3 increased as infection progresses, similar to histone dynamics observed in HSV-1 (Cliffe and Knipe, 2008).
Conclusion
In this study we show that MHV68 DNA undergoes a dynamic association with histone H3 during de novo infection of primary macrophages and MEFs. Many parameters of MHV68 chromatin are similar to those reported for HSV-1, but not HCMV, including MOI dependence of viral genome association with histone H3 (Cliffe and Knipe, 2008) and decreased proportion of viral DNA associated with histones following initiation of viral DNA synthesis (Cliffe and Knipe, 2008;Oh and Fraser, 2008). We found that MHV68 promoters and orilyt sequences acquired histone H3 shortly following viral entry, suggesting that the chromatization of viral promoters is likely to regulate early infection events, including initiation of viral gene transcription. Future studies should identify host and viral factors driving this rapid chromatization. Furthermore, we found that, similar to HSV-1 (Cliffe and Knipe, 2008), partial removal of histone H3 from the template MHV68 DNA occurred in the presence of cidofovir, a potent inhibitor of viral DNA synthesis and late gene expression. This suggests that an immediate early/early MHV68 gene is involved in this process, similar to the role ascribed to ICP0, an immediate early HSV-1 protein involved in histone removal from HSV-1 DNA (Cliffe and Knipe, 2008). In conclusion, our studies establish a foundation for future investigations of host and viral factors regulating gammaherpesvirus chromatin during de novo infection of primary cells.
Materials and Methods
Animals and primary cell cultures
C57BL/6J (BL6) mice were obtained from Jackson Laboratories (Bar Harbor, Maine). Mice were housed and bred in a specific-pathogen-free barrier facility in accordance with federal and institutional guidelines. All experimental manipulations of mice were approved by the Institutional Animal Care and Use Committee of the Medical College of Wisconsin. Bone marrow was harvested from mice between 7 and 10 weeks of age. Primary bone marrow macrophages and murine embryonic fibroblasts were generated and infected as previously described (Tarakanova et al., 2007).
Viral stock preparation
NIH 3T12 cells were infected with wild type MHV68 (WUMS clone, NCBI U97553) at MOI=0.05. At 7 days post infection cells and media were collected by freeze-thawing twice and centrifuged at 1000×g for 20m at 4°C to pellet cell debris. Supernatant was transferred to new tubes and centrifuged at 12,000×g for 2h at 4°C. Pellet was resuspended in buffer with 40 U DNase (Ambion, Austin, TX) and incubated at 37°C for 1h. DMEM containing 10% FBS was added following DNAse digestion and virus stock preparation centrifuged at 12,000×g for 2h. Pellet was resuspended in DMEM supplemented with 10% FBS, aliquoted, and stored at −80°C.
Virus infections and cidofovir treatment
Wildtype MHV68 was titered on NIH 3T12 cells as previously described (Weck et al., 1996). Bone marrow-derived macrophages were infected at the indicated multiplicities of infection for 1 hr at 37°C and 5% CO2 to allow for adsorption and washed twice with PBS prior to replenishment of medium. Viral DNA synthesis was inhibited by maintaining macrophages in 1 μg/ml cidofovir (Gilead, Foster City, CA) immediately following adsorption.
Chromatin Immunoprecipitation
Infected cells were cross-linked with 1% formaldehyde for 10 minutes at room temperature, and cross linking was quenched with 125mM glycine. Cells were washed three times with cold PBS, collected in 3 mL PBS by scraping, and pelleted at 4°C. Cell pellet was resuspended in 2 mL lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl pH 8.1) with Halt protease cocktail (Thermo Scientific, Rockford, IL) and sonicated to produce approximately 500-bp fragments (Cell Disruptor 185, Branson Sonifier, Danbury, CT). Sonicated samples were cleared of debris by centrifugation and portion of supernatant was set aside for analysis of input DNA. Chromatin was precleared with 80 μL protein G sepharose beads (Invitrogen, Carlsbad, CA) for 1h at 4°C. Precleared chromatin was diluted 10-fold in the immunoprecipitation buffer (0.01% SDS, 1.1% Triton-10X, 1.2 mM EDTA, 16.7 mM Tris-HCl pH 8.1, and 176 mM NaCl, 10 μL Halt protease inhibitor cocktail [Thermo Scientific, Waltham, MA]), immunoprecipitated with 2 μg anti-histone H3 (AbCam, Cambridge, MA) or rabbit IgG (Santa Cruz Biotechnology, Santa Cruz, CA) overnight, and incubated with 60 μL protein G for 2h at 4°C. Immunoprecipitates were washed at 4°C with buffer (0.1% SDS, 1% Triton X-100, 2mM EDTA, 20 mM Tris-HCl pH 8.1) containing low salt (150 mM NaCl) and high salt (500 mM NaCl). The samples were subsequently washed with LiCl buffer (0.25% LiCl, 1% IGEPAL-CA630, 1% sodium deoxycholate, 2 mM EDTA, 10 mM Tris-HCl pH 8.1) and twice with Tris-EDTA buffer (2 mM EDTA, 10 mM Tris-HCl pH 8.1). Chromatin was eluted twice by incubating immunoprecipitates with elution buffer (100 mM NaHCO3, 1% SDS, 10 mM DTT) at room temperature for 15 minutes. Cross links of immunoprecipitated and input samples were reversed by treatment with NaCl overnight at 65°C. Proteins were cleared by proteinase K treatment at 45°C for 1h, and DNA was purified using GeneJET PCR purification kit (Fermentas, Glen Burnie, MD). Gene-specific DNA was amplified utilizing 280 nM primers directed against GAPDH (Forward: 5′-TGT-GAT-GGG-TGT-GAA-CCA-CGA-GAA-3′; Reverse: 5′-GAG-CCC-TTC-CAC-AAT-GCC-AAA-GTT-3′), core gene 50 promoter (Forward: 5′-TTT-AGC-ATC-TGC-CCG-ACC-TGA-GA-3′; Reverse: 5′-AAT-GGA-CCT-TGA-AAC-CCG-TGA-AGG-3′), distal gene 50 promoter (Forward: 5′-AGG-TGG-TGT-TGG-GTT-AGT-ACA-GCA-3′; Reverse: 5′-TAG-TGA-CAG-GTA-AAG-CAT-AGC-CTG-GG-3′), origin of lytic replication (Forward: 5′-GTG-TGG-CCT-TTG-TGT-GCC-TGT-AAA-3′; Reverse: 5′-AAA-TCG-GTT-TGC-GGT-TAG-ACC-AGG-3′), and orf57 promoter primers (Forward: 5′-AGA-ACA-GCT-TCG-TGC-TGA-CAA-ACC-3′; Reverse: 5′-TTT-GGT-AAG-CTG-GCC-ACA-GTC-TTG-3′). Amplification of DNA sequences was performed with Bulldog Taq polymerase (Portsmouth, NH) in the presence of SYBR Green (Invitrogen, Carlsbad, CA). PCR conditions used were 95°C for 30s followed by 40 cycles of 95°C for 10s, 57°C for 30s, and 72°C for 30s, performed using BioRad iQ5. Amplification efficiency was determined for all primer sequences using serially-diluted DNA purified from virally-infected cells. Enrichment was calculated using the ΔΔCT method.
Viral DNA quantitation
Input samples from sheared chromatin were probed with primers directed against the core gene 50 promoter and cellular GAPDH and analyzed by quantitative RT-PCR. Relative viral DNA was calculated using ΔCT values comparing GAPDH and core gene 50 promoter sequences, normalizing to 0 hpi (Fig. 1) or 1 hpi (Fig. 2) or normalizing the value obtained at 30 hpi or 48 hpi with cidofovir treatment to 1 (Fig. 3C, D).
qRT-PCR quantitation of viral messages
Total RNA was harvested, DNAse treated, and reverse transcribed in the presence or absence of the enzyme as described in (Tarakanova et al., 2009). cDNA and corresponding minus RT reactions were serially diluted (8-fold) and assessed, in triplicate, by real time PCR using iCycler (Bio-Rad, Hercules, CA). Gene 50 specific cDNA was amplified using primers decribed in (Tarakanova et al., 2009). Delta Ct method was used to quantify fold difference in the levels of RTA sequences between plus and minus RT reactions.
Highlights.
Gammaherpesviral DNA associates with histone H3 in lytic infection of primary cells
Dynamics of histone association dependent on multiplicity of infection
Viral DNA synthesis regulates histone H3 association
Acknowledgments
Expert technical and managerial support was provided by Brittani Wood. We thank Amy Hudson’s, William Jackson’s, and Scott Terhune’s laboratories for helpful discussions.
This work was supported by F32DK079649-01 (S.K.), R01DK073641 (L.A.C), Advancing Healthier Wisconsin and 1R56AI084889 (V.L.T.). The funding sources had no involvement in the published studies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Ambagala AP, Bosma T, Ali MA, Poustovoitov M, Chen JJ, Gershon MD, Adams PD, Cohen JI. Varicella-zoster virus immediate-early 63 protein interacts with human antisilencing function 1 protein and alters its ability to bind histones h3.1 and h3.3. Journal of Virology. 2009;83:200–209. doi: 10.1128/JVI.00645-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliffe AR, Knipe DM. Herpes simplex virus ICP0 promotes both histone removal and acetylation on viral DNA during lytic infection. Journal of Virology. 2008;82:12030–12038. doi: 10.1128/JVI.01575-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen GH, Ponce dL, Diggelmann H, Lawrence WC, Vernon SK, Eisenberg RJ. Structural analysis of the capsid polypeptides of herpes simplex virus types 1 and 2. Journal of Virology. 1980;34:521–531. doi: 10.1128/jvi.34.2.521-531.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuevas-Bennett C, Shenk T. Dynamic histone H3 acetylation and methylation at human cytomegalovirus promoters during replication in fibroblasts. Journal of Virology. 2008;82:9525–9536. doi: 10.1128/JVI.00946-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H, Chu JT, Park NH, Sun R. Identification of cis sequences required for lytic DNA replication and packaging of murine gammaherpesvirus 68. J Virol. 2004;78:9123–9131. doi: 10.1128/JVI.78.17.9123-9131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efstathiou S, Ho YM, Hall S, Styles CJ, Scott SD, Gompels UA. Murine herpesvirus 68 is genetically related to the gammaherpesviruses Epstein-Barr virus and herpesvirus saimiri. Journal of General Virology. 1990;71:1365–1372. doi: 10.1099/0022-1317-71-6-1365. [DOI] [PubMed] [Google Scholar]
- Favre M, Breitburd F, Croissant O, Orth G. Chromatin-like structures obtained after alkaline disruption of bovine and human papillomaviruses. Journal of Virology. 1977;21:1205–1209. doi: 10.1128/jvi.21.3.1205-1209.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson W, Roizman B. Compartmentalization of spermine and spermidine in the herpes simplex virion. Proc Natl Acad Sci USA. 1971;68:2818–2821. doi: 10.1073/pnas.68.11.2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray KS, Allen RD, Farrell ML, Forrest JC, Speck SH. Alternatively initiated gene 50/RTA transcripts expressed during murine and human gammaherpesvirus reactivation from latency. Journal of Virology. 2008 doi: 10.1128/JVI.01444-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunther T, Grundhoff A. The epigenetic landscape of latent Kaposi sarcoma-associated herpesvirus genomes. PLoS Pathog. 2010;6:e1000935. doi: 10.1371/journal.ppat.1000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino Y, Katano H, Zou P, Hohman P, Marques A, Tyring SK, Follmann D, Cohen JI. Long-term administration of valacyclovir reduces the number of Epstein-Barr virus (EBV)-infected B cells but not the number of EBV DNA copies per B cell in healthy volunteers. Journal of Virology. 2009;83:11857–11861. doi: 10.1128/JVI.01005-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent JR, Zeng PY, Atanasiu D, Gardner J, Fraser NW, Berger SL. During lytic infection herpes simplex virus type 1 is associated with histones bearing modifications that correlate with active transcription. Journal of Virology. 2004;78:10178–10186. doi: 10.1128/JVI.78.18.10178-10186.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipe DM, Cliffe A. Chromatin control of herpes simplex virus lytic and latent infection. Nat Rev Microbiol. 2008;6:211–221. doi: 10.1038/nrmicro1794. [DOI] [PubMed] [Google Scholar]
- Kutluay SB, Triezenberg SJ. Regulation of histone deposition on the herpes simplex virus type 1 genome during lytic infection. Journal of Virology. 2009;83:5835–5845. doi: 10.1128/JVI.00219-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Collins CM, Mendel JB, Iwakoshi NN, Speck SH. Gammaherpesvirus-driven plasma cell differentiation regulates virus reactivation from latently infected B lymphocytes. PLoS Pathog. 2009;5:e1000677. doi: 10.1371/journal.ppat.1000677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman PM. Chromatin organization and virus gene expression. J Cell Physiol. 2008;216:295–302. doi: 10.1002/jcp.21421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Pavlova IV, Virgin HW, Speck SH. Characterization of gammaherpesvirus 68 gene 50 transcription. Journal of Virology. 2000;74:2029–2037. doi: 10.1128/jvi.74.4.2029-2037.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillen J, Consigli RA. In vitro radioisotopic labeling of proteins associated with purified polyoma virions. Journal of Virology. 1974;14:1627–1629. doi: 10.1128/jvi.14.6.1627-1629.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neyts J, De CE. In vitro and in vivo inhibition of murine gamma herpesvirus 68 replication by selected antiviral agents. Antimicrob Agents Chemother. 1998;42:170–172. doi: 10.1128/aac.42.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitzsche A, Paulus C, Nevels M. Temporal dynamics of cytomegalovirus chromatin assembly in productively infected human cells. Journal of Virology. 2008;82:11167–11180. doi: 10.1128/JVI.01218-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J, Fraser NW. Temporal association of the herpes simplex virus genome with histone proteins during a lytic infection. Journal of Virology. 2008;82:3530–3537. doi: 10.1128/JVI.00586-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulus C, Nitzsche A, Nevels M. Chromatinisation of herpesvirus genomes. Rev Med Virol. 2010;20:34–50. doi: 10.1002/rmv.632. [DOI] [PubMed] [Google Scholar]
- Peng H, Nogueira ML, Vogel JL, Kristie TM. Transcriptional coactivator HCF-1 couples the histone chaperone Asf1b to HSV-1 DNA replication components. Proc Natl Acad Sci USA. 2010;107:2461–2466. doi: 10.1073/pnas.0911128107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignatti PF, Cassai E. Analysis of herpes simplex virus nucleoprotein complexes extracted from infected cells. Journal of Virology. 1980;36:816–828. doi: 10.1128/jvi.36.3.816-828.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Placek BJ, Huang J, Kent JR, Dorsey J, Rice L, Fraser NW, Berger SL. The histone variant H3.3 regulates gene expression during lytic infection with herpes simplex virus type 1. Journal of Virology. 2009;83:1416–1421. doi: 10.1128/JVI.01276-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarakanova VL, Leung-Pineda V, Hwang S, Yang CW, Matatall K, Basson M, Sun R, Piwnica-Worms H, Sleckman BP, Virgin HW. Gamma-Herpesvirus Kinase Actively Initiates a DNA Damage Response by Inducing Phosphorylation of H2AX to Foster Viral Replication. Cell Host and Microbe. 2007;1:275–286. doi: 10.1016/j.chom.2007.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarakanova VL, Molleston JM, Goodwin M, Virgin HW., IV MHV68 complement regulatory protein facilitates MHV68 replication in primary macrophages in a complement independent manner. Virology. 2009 doi: 10.1016/j.virol.2009.10.030. Accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgin HW, Latreille P, Wamsley P, Hallsworth K, Weck KE, Dal Canto AJ, Speck SH. Complete sequence and genomic analysis of murine gammaherpesvirus 68. Journal of Virology. 1997;71:5894–5904. doi: 10.1128/jvi.71.8.5894-5904.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weck KE, Barkon ML, Yoo LI, Speck SH, Virgin HW. Mature B cells are required for acute splenic infection, but not for establishment of latency, by murine gammaherpesvirus 68. Journal of Virology. 1996;70:6775–6780. doi: 10.1128/jvi.70.10.6775-6780.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weck KE, Kim SS, Virgin HW, Speck SH. Macrophages are the major reservoir of latent murine gammaherpesvirus 68 in peritoneal cells. Journal of Virology. 1999;73:3273–3283. doi: 10.1128/jvi.73.4.3273-3283.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]