Fig. 2.
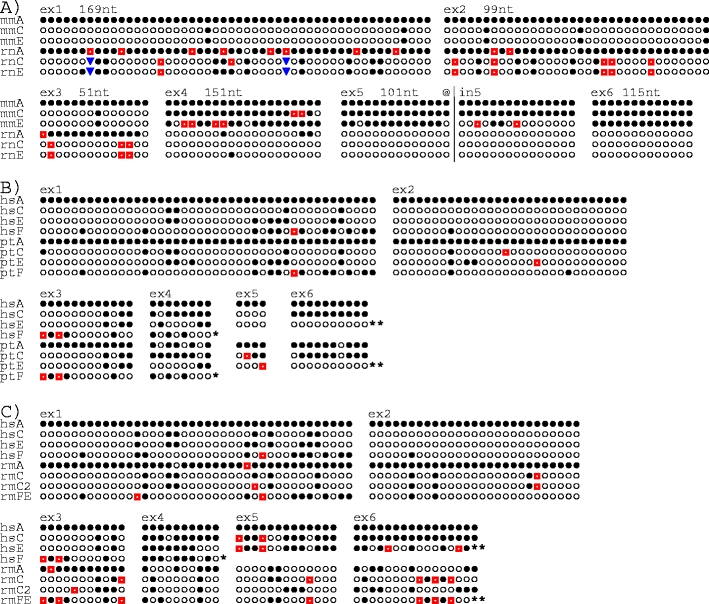
Exonwise comparisons of substitution sites in KLRC genes. a Mouse (mm) versus rat (rt), b human versus chimpanzee (pt), and c human (hs) versus rhesus monkey (rm). Nucleotide sequences for each exon were aligned and sites were removed if one of the following conditions was fulfilled: (1) all bases were identical, (2) only one of the genes differed from the others, (3) one or more of the genes exhibited gaps. The bases were replaced by symbols, where filled circles indicate the identity with the top sequence and the other symbols (open circles, red squares, blue inverted pyramids) the non-identical bases. Above the sequences are shown exon numbers, and in a also the numbers of compared bases. For example, 115 bases were compared for exon 6. At 92 sites, all six were identical and at ten sites only one of the six deviated from the others. These 102 sites were omitted, leaving the 13 sites shown. a The truncated exon 3 of mmNnkg2e shown in Fig. 1 (reported in Vance et al. 1998) may represent a splice variant; in the mouse genome sequence mmNkg2e was found to contain a sequence identical to exon 3 of mmNkg2c, as shown here. The alignment for exon 5 has been extended ~100 nt into intron 5 (@|in5) to demonstrate the changing affiliation of mmNkg2e (see “Results”). b The sequence referred to as ptC is based on the ptNKG2CI 01 allele (Khakoo et al. 2000). ptNKG2CII has, for clarity, been omitted. Single asterisk, hsNKG2F and ptNKG2F lack exons 5 and 6. b, c Two asterisks, as first described for hsNKG2E (Adamkiewicz et al. 1994) this gene as well as ptNKG2E and rmNKG2FE have insertion–deletion affecting the last ~20 nt of exon 6, resulting in sequence alteration of the conserved β5-strand, including the loss of the cysteine involved in disulphide bonding to the α1-helix. These sites are not shown
