Table 1. Crystallographic data and refinement statistics for the R2-state Hb complex structures.
Values in parentheses are for the outermost resolution bin.
| INN-298 | INN-312 | |
|---|---|---|
| Data-collection statistics | ||
| Space group | P3221 | P3221 |
| Unit-cell parameters (Å) | a = b = 92.0, c = 143.9 | a = b = 92.1, c = 143.8 |
| Molecules in asymmetric unit | 1 tetramer | 1 tetramer |
| Resolution (Å) | 38.75–1.80 (1.86–1.80) | 20.51–2.20 (2.28–2.20) |
| No. of measurements | 350101 (37076) | 167624 (16583) |
| Unique reflections | 65807 (6480) | 36347 (2797) |
| 〈I/σ(I)〉 | 14.6 (4.5) | 10.5 (3.3) |
| Completeness (%) | 99.9 (100) | 99.6 (100) |
| Rmerge† (%) | 5.9 (34.6) | 6.7 (34.2) |
| Structure refinement | ||
| Resolution limits (Å) | 27.78–1.80 (1.88–1.80) | 20.09–2.20 (2.30–2.20) |
| σ cutoff (F) | 0.0 | 0.0 |
| No. of reflections | 65805 (7700) | 36267 (4500) |
| R factor (%) | 20.1 (36.5) | 20.9 (26.9) |
| Rfree‡ (%) | 23.0 (38.0) | 26.8 (36.0) |
| R.m.s.d. from standard geometry | ||
| Bond lengths (Å) | 0.018 | 0.011 |
| Bond angles (°) | 2.1 | 1.8 |
| Dihedral angles | ||
| Most favored (%) | 92.0 | 91.4 |
| Allowed regions (%) | 7.6 | 8.4 |
| Average B factors (Å2) | ||
| All atoms | 32.3 | 46.4 |
| Protein | 31.4 | 45.8 |
| Heme | 27.4 | 43.7 |
| Water | 41.8 | 53.7 |
| INN | 37.1 | 47.4 |
R
merge = 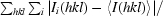
 .
.
R free was calculated with 5% of the reflections, which were excluded from the refinement.
