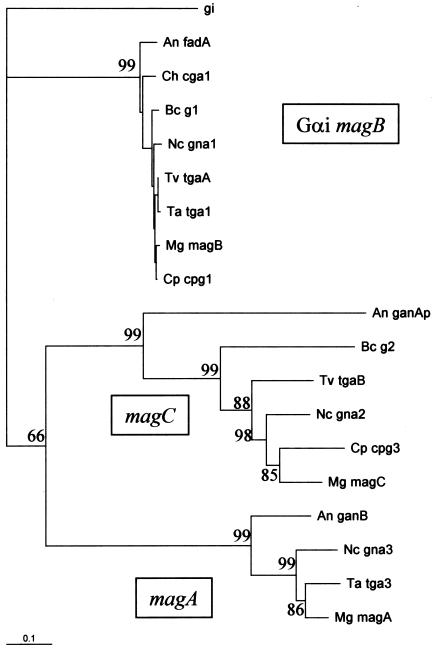
FIG. 1.
Phylogenetic relationships of tgaA and tgaB to other G-protein α subunits based on protein sequences. Binomial names are indicated by a two-letter code, followed by the gene name, as follows (the corresponding protein database accession numbers are also indicated): Mg, M. grisea (magB, O13315; magA, AAB65425; magC, AAB65427); Cp, Cryphonectria parasitica (cpg1, Q00580; cpg3, AAM14395); Nc, N. crassa (gna1, QO5425; gna2, Q05424; gna3, Q9HFW7); Ta, T. atroviride (tga1, AAK74191; tga3, AAM69919); Tv, T. virens (tgaA, AAO18659; tgaB, AAN65182); Bc, B. cinerea (G1, CAC19871; G2, CAC19872); Ch, C. heterostrophus (cga1, O74227); An, A. nidulans (fadA, Q00743; ganA, AAD34893; ganB, AAF12813). Gi, a human gene encoding Gαi subunit 2 (accession no. NP_006487). The scale bar indicates 0.1 nucleotide substitutions per site, and the numbers at the forks indicate the number of times the group consisting of the species which are to the right of that fork occurred among the trees, of 100 trees generated from the distance matrix. The groups have not yet been assigned standard names, so they are labeled here, arbitrarily, according to the nomenclature of M. grisea, a plant pathogen for which all three genes have been characterized.