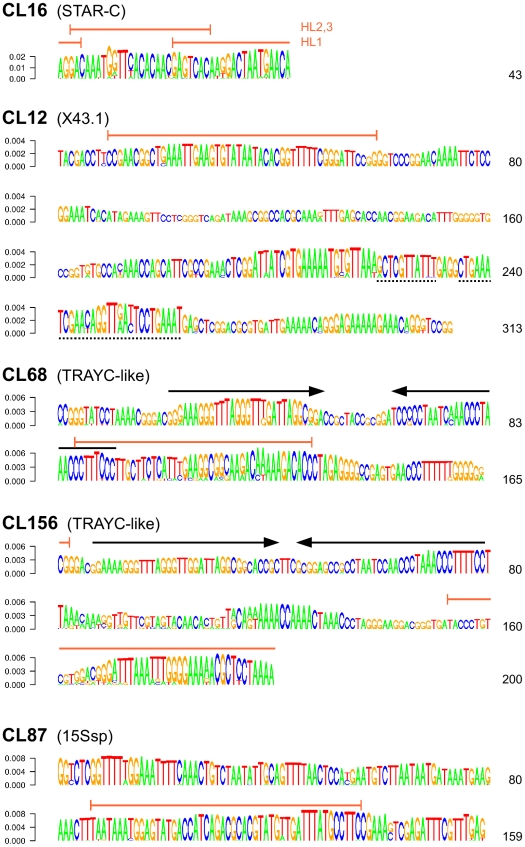
Figure 4. Consensus sequences of S. latifolia satellite repeats reconstructed from the most frequent k-mers.
K-mer frequencies for each satellite were calculated from Illumina sequencing data and used to reconstruct their most conserved fragments. These fragments were merged to create full-length monomers as depicted in Fig. S4. The resulting consensus sequences are displayed as sequence logos where the height of the letters reflects the frequencies of the corresponding k-mers, and major sequence variants are displayed along with the prevailing nucleotides [23]. The sequence regions used to design the FISH probes are marked with orange lines and the palindrome sequences common to the CL68 and CL156 repeats are marked with black arrows. The dashed underlines in the CL12 sequence show the positions of two regions that are duplicated in some of the X43.1 monomers.