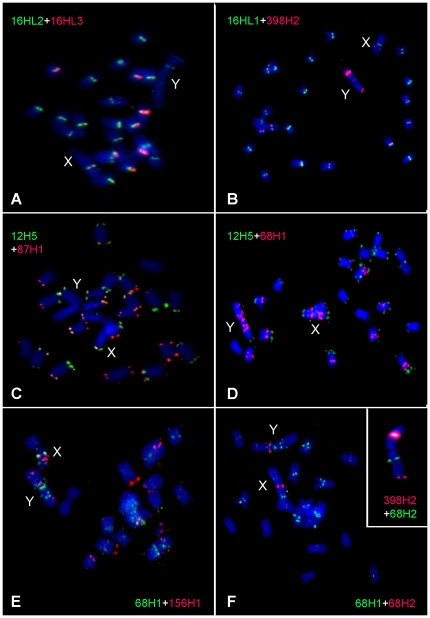
Figure 5. Localization of satellite repeats on metaphase chromosomes of S. latifolia.
FISH experiments were performed simultaneously with two probes labeled by different fluorochromes (red or green, as indicated) in order to investigate the co-localization of sequence variants of the STAR repeats (A–B), 15Ssp and X43.1 (C) and TRAYC-like repeats (D–F). (A) Co-localization of the CL16/STAR-C repeat consensus (16HL2) and its sequence variant (16HL3). (B) The probes for a different region of the CL16/STAR-C consensus (16HL1) and for the chromosome Y-enriched subfamily CL398/STAR-Y (398H2). (C) Consensus probes for CL87/15Ssp (87H1) and X43.1 (12H5) satellites. Co-localization of the CL68/TRAYC-like consensus (68H1) with X43.1 (12H5) (D), CL156 (156H1) (E) and with the CL68 subfamily (68H2) (F). The inset in (F) is an example of the Y chromosome hybridized to 68H2 and 398H2, showing the localization of their interstitial signals on different chromosome arms. The chromosomes were counterstained with DAPI (blue). Sex chromosomes are indicated with X and Y. The positions of probes within repeat monomers are shown on Fig. 4 and their sequences are provided in Table 2.