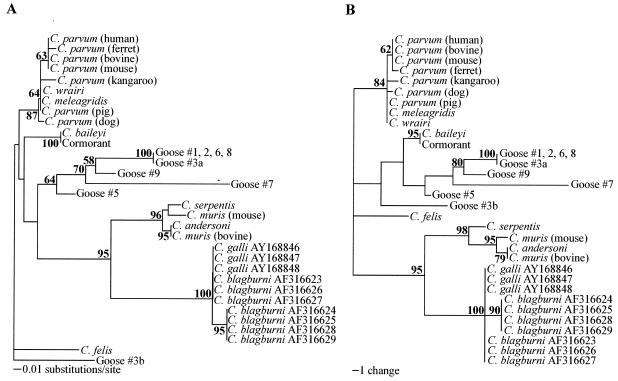
FIG. 3.
Phylogenetic analysis of the partial hypervariable region of the 18S rRNA gene to assess the relationships between the goose-derived sequences in the present study and C. galli and C. blagburni in finches. Shown are neighbor-joining (A) and most parsimonious (B) trees created with PAUP 4.0. C. felis was designated an outgroup. Evolutionary distances were determined by the Kimura two-parameter method. GenBank accession numbers of sequences included in the trees are AB089285 (C. andersoni), L19068 (C. baileyi), AF112575 (C. felis), AF112574 (C. meleagridis), L19069 (C. muris bovine genotype), AB089284 (C. muris murine genotype), AF093489 (C. parvum human genotype), AF093493 (C. parvum bovine genotype), AF112571 (C. parvum mouse genotype), AF112572 (C. parvum ferret genotype), AF115377 (C. parvum pig genotype), AF112576 (C. parvum dog genotype), AF112570 (C. parvum kangaroo genotype), AF093499 (C. serpentis), U11440 (C. wrairi), AY168846 through AY168848 (C. galli), AF316623 through AF316629 (C. blagburni), AY324634 (cormorant), AY324635 (goose no. 1), AY324636 (goose no. 2), AY324637 (goose no. 3 [sequence a]), AY324638 (goose no. 3 [sequence b]), AY324639 (goose no. 5), AY324640 (goose no. 6), AY324641 (goose no. 7), AY324642 (goose no. 8), and AY324643 (goose no. 9). Bootstrap values greater than 50% are indicated in bold at each respective node.