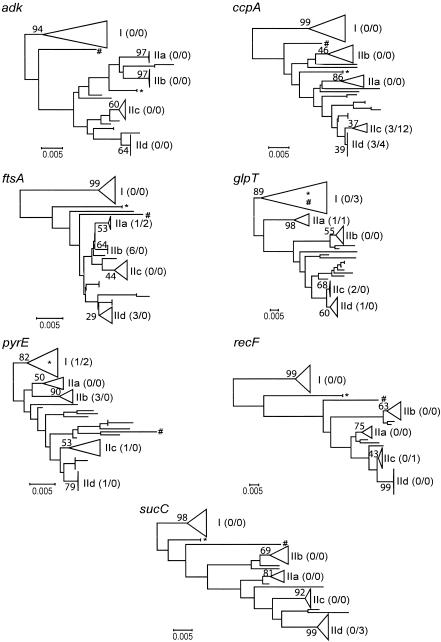
FIG. 2.
Phylogenetic relationships among 77 B. cereus group isolates inferred from individual genes. Trees were constructed by using the NJ method applied to pairwise distances among strains, computed by using Kimura's (19) nucleotide substitution model. Five groups of strains that were recovered for each tree were designated I, IIa, IIb, IIc, and IId. For the sake of simplicity, strain designations and detailed relationships among isolates within the groups are not shown (detailed trees and compositions of the clusters are available from us). For each of the five groups, the numbers in parentheses indicate the number of missing isolates/number of additional isolates compared to the number of isolates in the adk tree, and the numbers above the branches indicate bootstrap values, expressed as percentages (based on 1,000 replicates). In the adk tree, clusters I, IIa, IIb, IIc, and IId contain 18, 8, 12, 13, and 14 strains, respectively. B. anthracis belongs to cluster IIc. Isolates AH 1272 and AH 1273 (ST 52) and AH 1247 (ST 49), which cluster within cluster I, within cluster II, or between the two groups depending on the gene analyzed, are indicated by an asterisk and a number sign, respectively. The scale bars indicate 0.005 nucleotide substitution per site.