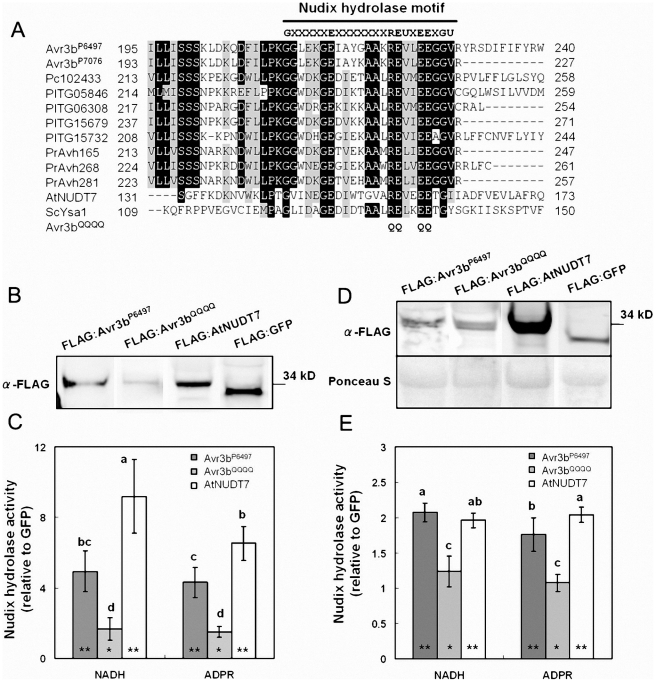
Figure 4. Avr3b is an ADP-ribose/NADH pyrophosphorylase.
(A) Sequence alignment of the Nudix motif region from Avr3b homologs and characterized nudix hydrolases. Pc102433 is a predicted P. capsici RXLR effector. PITG05846, PITG06308, PITG15679, and PITG15732 are predicted P. infestans RXLR effectors. PrAvh165, PrAvh268, and PrAvh281 are P. ramorum RXLR effectors. AtNUDT7 is an ADP-ribose/NADH pyrophosphorylase from Arabidopsis. ScYSA1 is an ADP-ribose pyrophosphorylase from Saccharomyces cerevisiae. The black line refers to the Nudix hydrolase motif “G5XE7XREUXEEXGU.” The threshold for amino acid shading in this alignment is 70%. Avr3bQQQQ is an Avr3bP6497 non-functional mutant; the glutamine (Q) substitutions in the nudix hydrolase motif are marked. (B) Western blot of purified Avr3b and control proteins. Immuno-precipitated recombinant proteins FLAG:AtNUDT7, FLAG:GFP, FLAG:Avr3bP6497 and predicted non-functional Avr3b mutant FLAG:Avr3bQQQQ were detected with anti-FLAG antibody. (C) Enzyme assays of purified Avr3b and control proteins. The hydrolase activities of FLAG:AtNUDT7, FLAG:Avr3bP6497 and FLAG:Avr3bQQQQ were measured separately with FLAG:GFP as a control for each assay. The relative hydrolase activity was calculated as a ratio of Avr3b/AtNUDT7 over GFP. ADPR: adenosine diphosphate ribose, NADH: nicotinamide adenine dinucleotide (reduced form). Bars represent standard errors from 6 independent replicates each. Student's t-test was used to compare whether testing sample activities are significantly different from the GFP control. The double asterisk and single asterisk indicates statistical significance p <0.01 and p <0.05, respectively. The letters represent statistical significance (P <0.01) as measured by Duncan's multiple range test. (D) Western blot of the total plant extract from the Agrobacterium infiltrated region. The supernatants from FLAG:AtNUDT7, FLAG:GFP, FLAG:Avr3bP6497 and FLAG:Avr3bQQQQ transgenic N. benthamiana leaves were electrophoresed and detected with anti-FLAG antibody. (E) Enzyme assays of the total protein extracts from the Agrobacterium infiltrated region. The relative hydrolase activities were measured as a ratio to extracts from a GFP-expression control. Bars represent standard errors from 4 independent replicates each. Student's t-test was used to compare whether testing sample activities are significantly different from the GFP control. The double asterisk and single asterisk indicates statistical significance p <0.01 and p <0.05, respectively. The letters represent statistical significance (P <0.01) as measured by Duncan's multiple range test.