Table 2. Data extracted from the MD simulations.
| 12-10-12 | 12-2-12 | 14-2-14 | 18-2-18 | |
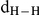 /nm /nm |
1.4±0.2 | 0.5±0.1 | 0.5±0.1 | 0.5±0.1 |
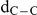 /nm /nm |
1.9±0.7 | 1.0±0.6 | 0.9±0.4 | 0.9±0.4 |
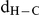 /nm /nm |
1.2±0.1 | 1.2±0.1 | 1.3±0.1 | 1.7±0.1 |
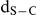 /nm /nm |
0.9±0.3 | 1.2±0.1 | 1.3±0.2 | 1.7±0.2 |
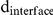 /nm /nm |
0.49±0.07 | 0.21±0.02 | 0.20±0.04 | 0.09±0.03 |
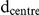 /nm /nm |
0.21±0.08 | 0.36±0.04 | 0.32±0.05 | -0.12±0.04 |
General data on the average conformation (top of the table) and relative positioning (bottom of the table) of the indicated gemini molecules inserted in a fully hydrated DPPC bilayer as extracted from the MD simulation at 325 K. See Figure 8 for a schema of the respective distances.
