Figure 2. Apparent tertiary structure similarity between various solved crystal structures used in this study.
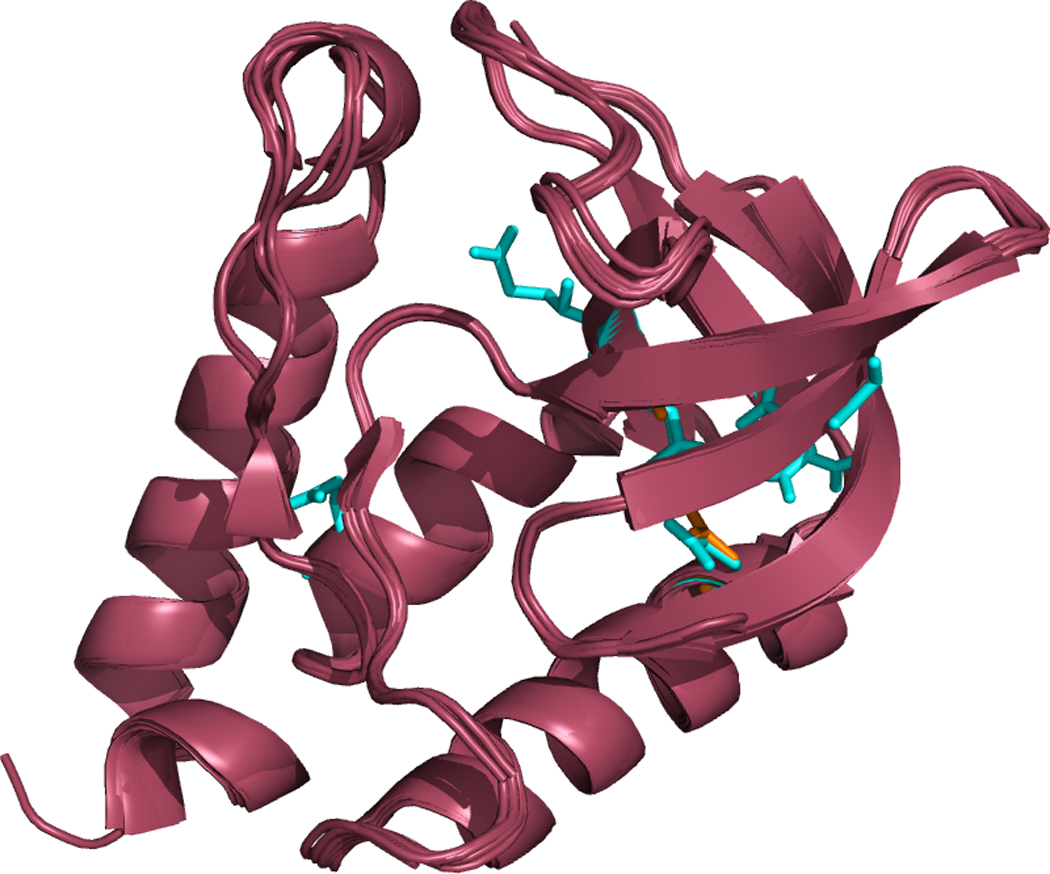
Δ+PHS staphylococcal nuclease, its 6 solved PDB structures, and three structural homologues are all shown overlaid with one another. The mutated residues are shown in red. All mutants had an RMSD of less than 0.35 Å, indicating that even with the introduction of hydrophilic residues into the protein’s interior, the structure of Δ+PHS is not significantly distorted.
