Abstract
Deleterious mutations tend to be recessive. Several theories, notably those of Fisher (based on selection) and Wright (based on metabolism), have been put forward to explain this pattern. Despite a long-lasting debate, the matter remains unresolved. This debate has focused on the average dominance of mutations. However, we also know very little about the distribution of dominance coefficients among mutations, and about its variation across environments. In this article we present a new approach to predicting this distribution. Our approach is based on a phenotypic fitness landscape model. First, we show that under a very broad range of conditions (and environments), the average dominance of mutation of small effects should be approximately one-quarter as long as adaptation of organisms to their environment can be well described by stabilizing selection on an arbitrary set of phenotypic traits. Second, the theory allows predicting the whole distribution of dominance coefficients among mutants. Because it provides quantitative rather than qualitative predictions, this theory can be directly compared to data. We found that its prediction on mean dominance (average dominance close to 0.25) agreed well with the data, based on a meta-analysis of dominance data for mildly deleterious mutations. However, a simple landscape model does not account for the dominance of mutations of large effects and we provide possible extension of the theory for this class of mutations. Because dominance is a central parameter for evolutionary theory, and because these predictions are quantitative, they set the stage for a wide range of applications and further empirical tests.
DOMINANCE plays a prominent role in evolution. For instance the genetic load with inbreeding (e.g., Whitlock 2002), the magnitude of inbreeding depression (e.g., Bataillon and Kirkpatrick 2000; Charlesworth and Willis 2009), the rate of adaptation in diploids (e.g., Orr and Otto 1994), the evolution of mating systems (e.g., Epinat and Lenormand 2009), dispersal (e.g., Roze and Rousset 2005), life cycles (e.g., Otto and Goldstein 1992), polyploid tissues (e.g., Cailleau et al. 2010), and sex (e.g., Agrawal 2009) depend on the distribution of dominance of mutations, to cite a few; there are other situations where dominance matters (Charlesworth and Charlesworth 1998; Lynch et al. 1999).
It is possible to precisely define the dominance of a mutation on any particular character, by expressing the character value of the heterozygote as a fraction of the difference in character values between the two homozygotes. Thus, dominance is not the absolute property of a mutation, which may be dominant on a character and recessive on another (Bürger and Bagheri 2008) or have varying dominance in different environments (Bourguet et al. 1996). In this article, the character of interest is fitness, the dominance of which “became one of the longest and fiercest controversies in the history of evolutionary biology” (Orr 1991). This debate even “marked the end of [the] previously cordial interchange of ideas” between Wright and Fisher (Provine 1986). In Fisher’s theory (1928), the repeated occurrence of deleterious mutations should select for more dominant “wild-type” alleles by modifier genes, leading to genotypes increasingly sheltered against the harmful effects of mutation. Three problems with this explanation have been raised. First, the selection pressure in favor of a modifier of dominance is very weak, proportional to the mutation rate: it could thus be easily overwhelmed by any deleterious pleiotropic effect of dominance modifiers. This was essentially the objection of Wright (1929, 1934). Although this may not be a problem in a very large population or if mutations are maintained polymorphic at high frequency (e.g., by migration; Otto and Bourguet 1999), it clearly limits the scope of this theory. Second, the selection pressure in favor of a modifier of dominance should depend on its effect on dominance but not on the strength of selection, which was contradicted by the fact that lethal mutations are more recessive than nonlethal ones in Drosophila (Charlesworth 1979). Third, mutations in Chlamydomonas, an almost exclusively haploid organism, also tend to be recessive, even when considering traits that are uniquely expressed in the haploid stage (such as their flagella). Because there has been no opportunity to select for dominance in such a haploid organism, this finding was also claimed to falsify Fisher’s theory (Orr 1991, but see below).
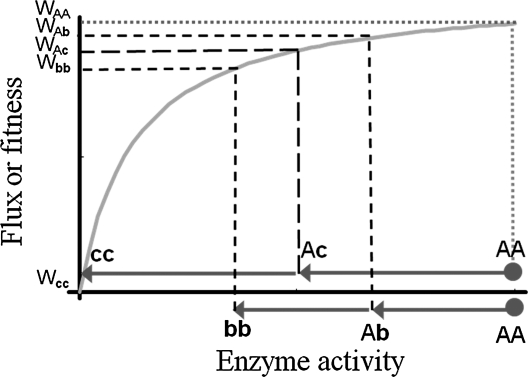
In Wright’s theory (Wright 1934), later elaborated as the “metabolic control theory” (Kacser and Burns 1981), recessivity results from the fact that enzymes in a metabolic pathway must share the control of the flux through this pathway (although this argument may be weaker in complex pathways; Omholt et al. 2000; Bagheri and Wagner 2004). As a consequence, the flux or concentration of metabolites in a pathway can change substantially only if one enzyme’s activity is dramatically reduced (Figure 1). Thus, provided fitness is a linear function of the flux or metabolite concentration, the recessivity of deleterious mutations naturally emerges. Over the years, these ideas have been much refined, but the pivotal role of metabolic biochemistry to account for dominance is still controversial (De Visser et al. 2003; Bagheri and Wagner 2004; Bürger and Bagheri 2008). Nevertheless, this theory has a wide appeal, in particular because it is rooted in a mechanistic explanation for the robustness of enzymatic pathways, and because it avoids the issues raised against Fisher’s theory (Bourguet 1999). What are the main problems with this theory? First, it rests on enzymatic kinetics within cells. It ignores mutations that do not affect enzymes (Phadnis and Fry 2005); plus it ignores processes operating beyond cell level. For instance, the fitness consequences of mutations affecting how enzymes are regulated or where and when enzymes are expressed cannot be summarized by the variation in their kinetic constants. Second, and more importantly, metabolic theory of dominance implicitly assumes that fitness is linearly and positively related to flux in pathways [or similarly, linearly and positively (resp. negatively) related to downstream (resp. upstream) substrate concentrations in pathways)]. This assumption cannot be general because it excludes the possibility that intermediate flux or substrate concentrations would be optimal (Hurst and Randerson 2000). For instance it ignores the fitness cost of producing the enzymes and more generally trade-offs among resource use. It is important to stress here that not only monotonicity between metabolic flux and fitness is required, but linearity. While this may occur in some circumstances (Dykhuizen et al. 1987), it is far from being empirically confirmed in general. Third it is extremely difficult to predict overdominance with this theory. Overdominance is not something very common in nature (Gemmell and Slate 2006). However, even if it is rare, it is incompatible with a simple metabolic view of dominance. Finally, the same data that have been used to falsify Fisher’s theory can also be used to falsify the metabolic theory. Lethal mutations are more recessive than nonlethal mutations, but surprisingly, both have undistinguishable fitness effect when heterozygous in Drosophila (Simmons and Crow 1977) and yeast (Szafraniec et al. 2003). With a diminishing return curve relating enzymatic activity to fitness, as assumed in metabolic theory, it is straightforward to show that, in the heterozygous state, lethal mutations, instead, should be on average more deleterious than nonlethal mutations (Figure 1).
Figure 1.
Dominance of mutations as predicted by the metabolic control theory. A loss-of-function mutation (c) reducing enzymatic activity to 50% (resp. 0%) when heterozygous (resp. homozygous) has a lethal but strongly recessive effect on metabolic flux (or fitness). A mutation decreasing less drastically the activity of the enzyme (b) has a milder and less recessive effect on metabolic flux (or fitness). However, by construction (i) the heterozygous effect of the lethal mutation is larger than that of the nonlethal (WAb > WAc) and (ii) overdominance cannot occur.
The debate over dominance has been extremely polarized between the views of Fisher and Wright, with little space for alternative theories to be evaluated. One central reason for such a long-lasting debate has been the lack of clear and quantitative predictions on the distribution of dominance that could be confronted with data. Further, beyond the qualitative predictions made by the existing theories, it would be useful to provide quantitative predictions to really confront data and to develop more realistic evolutionary models for diploid organisms. The first aim of this article is to develop an alternative theory for the dominance of mutation fitness effects. The second aim is to show that this theory makes predictions that are upheld when confronted with the available data.
Our theory proposes that the recessivity of deleterious mutations is a necessary consequence of stabilizing selection for a given optimum on a set of (n) unknown phenotypic traits. These traits are not necessarily metabolic, but encompass life history, behavior, etc. This theory extends a model originally put forward by Fisher in another context (to defend his micromutationist view of evolution)—his “geometrical model of adaptation.” This model was not discussed in the context of dominance of mutations, but provides a general, yet unexplored, framework to predict it. Oddly enough, Wright was close to such an approach but did not develop it (Wright 1935). It is often viewed as heuristic, but it in fact provides a “top-down” approach that has few underlying assumptions, all of them being more realistic than often realized (Martin and Lenormand 2006b). Furthermore, this model has proved useful to quantitatively predict, for example, the distribution of epistasis among random mutations in two microbe species, following principles similar to this study (Martin et al. 2007). This approach also offers a quantitative prediction for the overall distribution of dominance, across distinct environments, which allows more powerful tests of the theory. We will see that these predictions are upheld when confronted with the available data, although further quantitative tests would be useful, and are possible with the experimental methods available today.
Model
Natural selection is often thought of as favouring one extreme of each measurable character. Doubtless there is such a process under exceptional conditions, but it is certainly more often the case that the best adapted individuals are those nearest the average in every respect. (Wright 1935)
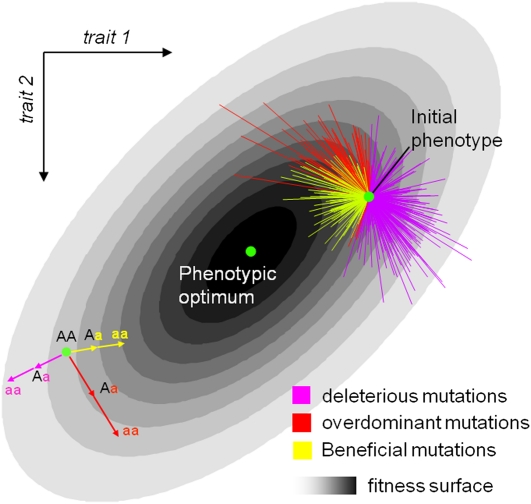
We used a generalization of Fisher’s model accounting for arbitrary mutational correlations and selective interactions between traits determining fitness. Any continuous model of stabilizing selection, including the present one, naturally generates dominance for fitness (W), even when mutations are additive on the underlying phenotype (z), because the relationship between phenotype and fitness can be nonlinear only close to the optimum (Figure 2). In fact, the same argument would hold for any secondary trait (not only fitness) that can be thought of as the optimization of a set of underlying primary traits (Wright 1935). Specifically, we use a Gaussian fitness function W(z) relating phenotype to fitness, which approximates any smooth function in the vicinity of an optimum (Lande 1976). The assumption of a single optimum is not very restrictive as it is relative to the scale of mutational variation: it requires only that the mutant cloud “sees” only a single optimum. We then assume that the distribution of the homozygous phenotypic effects of mutations around a wild-type phenotype is an arbitrary multivariate Gaussian with mean zero. This Gaussian assumption is not very restrictive either since the definition of each trait is arbitrary and may be chosen or transformed appropriately (Lande 1976). Finally, we model the phenotype of a heterozygote as a fraction υ of the phenotypic displacement corresponding to the mutant homozygote. In the additive case, the displacement for the heterozygote is simply half that of the homozygote. This approach is consistent with the fact that many metric characters are codominant. In a more general model υ is variable among mutants and additive only on average (E(υ) = 1/2, Var(υ) = συ2). This assumption is much less restrictive than the purely additive model. From this model, we can derive the joint distribution of dominance and selection coefficients and different measures of average dominance.
Figure 2.
Two-dimensional illustration of the mutation model. This figure illustrates the principle of the model in two dimensions (two traits). The fitness function is a bivariate Gaussian function, which increases toward an optimum with darker gray shading. Mutants are distributed around an initial phenotype or wild type (green spot). The bottom-left sketch explains how dominance on fitness arises even when the phenotype of the heterozygous mutant is halfway between the initial and the homozygous phenotype (additive phenotypes). The top-right sketch shows how a random draw of mutants around an initial phenotype (green spot) generates a distribution of homozygote and heterozygote fitness effects. Any model of stabilizing selection naturally generates dominance for fitness even when mutations act additively on the underlying phenotype. This is due to the concavity of the fitness surface around the optimum. With such a surface, deleterious mutations (purple) tend to be recessive because if a step in one direction is deleterious (i.e., it increases the distance to the optimum), two steps in the same direction necessarily result in more than twice the deleterious effect. The reverse is true for beneficial mutations (yellow) that tend to be dominant. Interestingly, this model also predicts that some mutations (red) should exhibit overdominance in their fitness effect if mutations that bring the phenotype close to the optimum in one step overshoot it in two steps in the same direction.
There are several ways to define dominance of a given mutation, the most frequent being h = (whet − 1)/(whom − 1) ≈ shet/shom. This well-known definition (corresponding to the fitness notation 1, 1 − h shom, 1 − shom) is undefined when shom = 0, so that no meaningful distribution of h can be obtained when shom encompasses positive and negative values. Alternatively, dominance of a given mutation may be defined as a departure from multiplicative fitness effects (Otto 2003)
| (1) |
The latter definition is mathematically more robust and can be used to derive a distribution of dominance in all cases. From an empirical point of view, an estimation of E(ι) may be conveniently used as a measure of average dominance. However, other approaches are most commonly used. Average dominance is often measured over a set of mutations, as either the ratio
| (2) |
or the slope (hR) of the regression of shet on shom computed from the conditional expectation
| (3) |
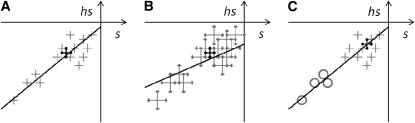
(shet0 being the intercept of the regression). Because of their definitions (hR as a slope of a regression and hM as the ratio of means), estimation of these two quantities poses different problems in presence of measurement error (noise) and of missing or omitted data. hM estimates are likely to be robust with respect to noise, but strongly biased by the omission of extreme points. hR estimates are likely to be robust to missing or omitted values, but downwardly and moderately biased by measurement error (we illustrate those behaviors on Figure 3).
Figure 3.
Sketch of the behavior of hR (line) and hM (thick cross) to measurement of noise and missing or omitted data. (A) All data, thick cross, ratio of means; line, regression. (B) Noisier data, the regression slope decreases, the ratio of means is less affected. (C) Missing or omitted large effects (open circles); the ratio of means changes, the regression slope is less affected.
Analysis
Here, we analyze the general n-dimensional model. As we see, accounting for dimension affects some, but not all results. However, to provide intuitive insights, we give in supporting information, File S1, the derivation in the simpler one-dimensional subcase. We note W(z) the fitness of a phenotype z (of dimension n, the number of phenotypic traits under selection). As explained above, W is multivariate Gaussian in our model
| (4) |
where superscript t denotes transposition, and S is an arbitrary n × n symmetric positive semidefinite matrix that describes all the selective interactions between phenotypic traits. We focus on a wild-type phenotype zo, whose fitness distance to the optimum is measured by so ≡ − log W(zo) = zotSzo. We then note zo + dzi the phenotype of a mutant homozygous for a mutation i (note that adding independent environmental noise contributing to the phenotype does not change the argument; Martin et al. 2007; Chevin et al. 2010). Its log-relative fitness shom is thus
| (5) |
The phenotypic effect of heterozygotes is determined by a fraction υ of the homozygote phenotypic displacement. Thus, the phenotype of the mutant heterozygote is zo + υ dzi. The log-relative fitness of the mutant heterozygote shet is
| (6) |
Defining dominance ι as a departure from multiplicative fitness effect (Equation 1), we find
| (7) |
Because dz has a multivariate Gaussian distribution in our model (with mean 0 and arbitrary positive semidefinite covariance matrix M), there is a linear transformation of the original phenotypic space such that M becomes the identity matrix I and the arbitrary matrix S becomes a diagonal matrix Λ (with diagonals elements λi equal to the eigenvalues of SM) (Martin and Lenormand 2006b). In this new base the phenotypic vector z becomes the vector x. With these new notations, Equation 7 becomes
| (8) |
Measures of average dominance
As explained above, there are different ways to measure average dominance. Mathematically, the most straightforward is to compute moments of ι from Equation 8. Its mean and variance are
| (9) |
| (10) |
where Tr(.) denotes matrix trace. Both depend on συ2 and Var(ι) also depends on E(υ4). To get a better intuitive sense of these results, it is useful to express them in terms of the mean and variance of homozygous fitness effects. To simplify Var(ι), it is also convenient to consider the case where υ is drawn in a symmetric β-distribution (a very flexible assumption encompassing uniform to approximately normal distribution of υ). We obtain
| (11) |
| (12) |
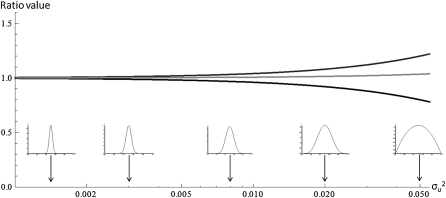
where ε = so/|E(shom)| measures the level of maladaptation of the wild type in a given environment and Var(shom) stands for the variance of shom when the wild type is at the optimum (ε = so = 0). Results (11) and (12) indicate that departures from a purely additive model of phenotypic effects of mutations has a very limited impact on the prediction of average dominance, since συ2 is likely to remain small (e.g., it is only ∼0.08 in the extreme case of a uniform distribution of υ). This result is illustrated on Figure 4. Moreover, Equations 11–12 also provide a direct method to estimate συ2 and check whether the data support nonadditive phenotypic effects. More usual measures of average dominance hM (Equation 2) or hR (Equation 3) yield
| (13) |
Figure 4.
Limited impact of departures from a purely additive model. Ratios average dominance computed with different values of συ2, relative to the value computed in the additive case. For illustration, the distribution of υ corresponding to different values of συ2 (cases συ2 = 0.0012, 0.003, 0.008, 0.02, 0.05 illustrated) are shown in the insets (the distribution are within [0,1]). Bottom line: ratio E(ι)/E(ι)add. Top line: ratios of hM/hM add and hR/hR add when the wild type is as optimum (so = 0) (they superpose). Gray line: ratio hR/hR add when the wild type is not at the optimum (case considered, so = 0.06 and E(shom) = 0.05).
If the phenotypic effect of mutations is additive (E(υ) = 1/2 and συ = 0) and the wild type is close to the phenotypic optimum (ε ≈ 0), then hM ≈ hR ≈ 1/4. This result is easily seen from Equation 5–6, noting that E(υ2) = . If the phenotypic effect of mutations is only additive on average (E(υ) = 1/2 but συ > 0), it tends to bias upward the average dominance (both hM and hR). However, this bias is not large. For instance, if we consider the extreme situation in which υ is uniformly distributed over the range [0, 1] (so that (συ = 1/12), we obtain, close to the optimum, hM ≈ hR ≈ 1/3. The distance to the optimum does not affect hM, whereas hR tends to 1/2 when the wild type is far away from the optimum. Note that when the wild type is not at the optimum, some mutations are beneficial (and tend to be dominant), and others are overdominant (because shet0 > 0 so that shet can be, e.g., positive while shom is negative; Figure 2).
Simulation results presented on Figure 5 (dots) were obtained by drawing both S and M matrices into a standard Wishart distribution W50[I,50] (a probability distribution for nonnegative-definite random matrices, in this case of dimension 50) and scaled so that in all cases E(shom) = 0.05. In Figure 5, the phenotypic effects (dzi) of one thousand mutations were sampled from a multivariate Gaussian distribution with a variance–covariance matrix M. In the top of Figure 5, the heterozygous phenotype is halfway between the homozygotes (υ = 1/2) whereas in the bottom, the phenotypic change in the heterozygote is computed as a random fraction υ of the homozygous phenotypic change with E(υ) = 1/2 and συ2 = 0.02 (more specifically, in the bottom, υ ∼ Beta[23/4, 23/4]). These simulations show the agreement with the analytical results described above. They also illustrate the dispersion of individual mutational effects under the different assumptions. Figure 6 gives hM for the different class of mutations (deleterious, overdominant with shom > 0, overdominant with shom < 0, beneficial) as a function of the scaled distance to the optimum ε. Despite the fact that each class-specific hM varies with ε, the hM averaged over all mutations stays constant with increasing distance to the optimum because the proportion of the different classes of mutations also vary with ε.
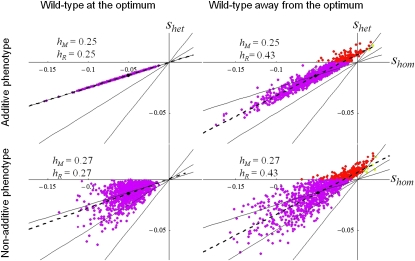
Figure 5.
Heterozygous vs. homozygous fitness effects under different models assumptions. The relationship between homozygous (shom, x-axis) and heterozygous (shet, y-axis) fitness effects of mutations is compared in four situations. Left vs. right: wild type is either at the optimum (so = 0, left) or away from it (so = 0.06, right). Top vs. bottom: mutation effects on the underlying phenotype are either all additive (top, E(ν) = 1/2, συ = 0) or only additive on average, with a random dominance coefficient (bottom, E(ν) = 1/2, συ2 = 0.02, 95% of υ values fall in the range [0.26–0.74]). Dots show the value of (shom, shet) for exact simulations of 1000 mutants, with corresponding values of hM and hR given on each graph. The same color code as in Figure 2 is used: purple, deleterious recessive; yellow, dominant beneficial; red, overdominant. The dashed line is the regression of shet on shom, which is undistinguishable from our prediction (hR and shet0, Equation 13). The black dot gives E(shet) and E(shom). The three plain lines indicate shet = shom/4, shet = shom/2 and shet = shom. hM varies little overall, but hR increases from 1/4 (left) to a value closer to 1/2 (right) when the wild type is far away from the optimum. In the latter situation, overdominant mutations and dominant beneficial mutations are also frequent.
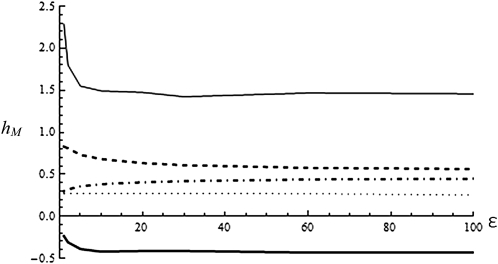
Figure 6.
Average dominance for different mutational classes. hM (y-axis) measured as a function of the scaled distance to the optimum ε. The dotted line (∙∙∙) is the average over all mutation and is constant. The dot-dashed (– . –) line indicates hM for mutations that are deleterious in both heterozygous and homozygous state (purple points on Figure 5). The dashed line (– – –) indicates hM for mutations that are beneficial in both heterozygous and homozygous state (yellow points on Figure 5). The solid line indicates hM for mutations that are overdominant, with beneficial (thin line) or deleterious (thick line) homozygous effect (red points on Figure 5). Values obtained by exact simulations with E(ν) = 1/2, συ2 = 0.02. Different values of E(shom) give undistinguishable curves.
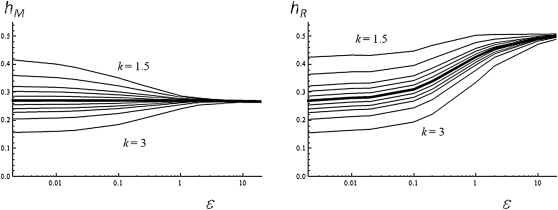
We also used simulation to investigate the effect of a departure from quadratic log-fitness on measures of average dominance. A different exponent (k) can be used in the fitness function to reflect this variation in peakedness (Martin and Lenormand 2006a; Tenaillon et al. 2007). We investigated k in the range 1.5–3 (more extreme k being very unlikely given the available evidence; Martin and Lenormand 2006a). Variation in k does affect average dominance, although the effect is limited in the most likely (narrower) range: hM ranges between 0.2 and 0.3 when k lies within [1.8, 2.3]. Interestingly, the departure from the k = 2 case also quickly decreases with increased distance to the optimum, of the order of one mutational step or more (i.e., at ε > 1; see Figure 7).
Figure 7.
Peakedness of the fitness function. Effect of the peakedness of the fitness function (k, as described in the text) on average dominance (hM, left; hR, right) as a function of the scaled distance to the optimum ε = so/E(shom). The case of a quadratic log-fitness (k = 2) is indicated by the thick line (other k values are from top to bottom: k = 1.5; 1.7; 1.8; 1.9; 2; 2.1; 2.2; 2.3; 2.5; 3). Average measure obtained by simulations (E(shom) = 0.05; συ2 = 0.02).
Distribution of dominance
As just mentioned, there is a large heterogeneity in the dominance of individual mutations in the model. This is readily seen in Equations 10 and 12 giving Var(ι) above. This heterogeneity is also large for h, the usual single mutation-based measure of dominance. In fact, for a given average dominance, individual mutations have a variety of dominance effects in the model, including overdominant mutations (h < 0 or h > 1) and dominant beneficial mutations (0.5 < h < 1) when the wild type is not at the optimum. Figure 2 illustrates this behavior and provides an intuitive explanation of this result. Overdominant mutations are mutations “overshooting” the optimum, where the heterozygote phenotype lies closer to the optimum than the phenotype of either homozygote. Beneficial mutations are the mutation pointing toward the optimum. They tend to be dominant because the fitness surface flattens near the optimum, yielding a diminishing return on fitness of the larger phenotypic change in homozygotes. We now focus on the full distribution of ι. Because departures from a purely additive model have a very limited impact on our predictions (see above), we derive below for simplicity the results under the additive assumption (υ = 1/2 exactly for all mutations). In this case, the distribution of homozygous effects shom reads
| (14) |
with mean and variance equal to
| (15) |
Similarly, the dominance ι now reads
| (16) |
The expressions (9) and (10) simplify to
| (17) |
From (16), we see that ι is a quadratic form in Gaussian vectors. Matching the two first moments in (17), it can be well approximated by a negative Gamma distribution of shape and scale parameters aι and bι. These parameters can be expressed in terms of measurable quantities:
| (18) |
| (19) |
Importantly, this result indicates that, assuming additivity on traits, the distribution of ι can be fully predicted using quantities measured from homozygous mutant fitnesses alone (E(shom), Var*(shom)). Note that while Equations 18–19 depend on the variance Var*(shom) in homozygous fitness effects when the wild type is at the optimum, this can still be computed using (15) when the wild type is not at the optimum, provided that ε can be estimated empirically.
To summarize, the dominance ι follows a negative Gamma distribution (−ι ∼ ) whose parameters a* and b* are those of the Gamma distribution of shom at the optimum (so = 0). These parameters can be estimated independently from data on homozygous effects alone; i.e., this model entirely predicts the distribution of ι without using any data on ι. The lack of a large and reliable data set on single mutants prevented us from confronting this prediction, but such a test would be a further way to challenge this theory.
Joint distribution of shom and ι
From Equation 14 and 16 we can see that shom and ι are not independent. Thus additional information and further tests of our theory may be obtained by studying their bivariate distribution. We derive this bivariate distribution assuming additivity on traits (υ = 1/2) as above. First we take the equivalent landscape, , where I has dimension ne = 2a* (Martin and Lenormand 2006b) to obtain spherical symmetry. We then express dx in cylindrical coordinates, in terms of its norm r (r2 = dx.dx) and θ the angle with respect to the direction of the optimum. We have
| (20) |
where the probability density function of y is
| (21) |
where denotes Euler’s Gamma function (Welch and Waxman 2003). This distribution is independent of r. It could be well approximated by a standard Gaussian as long as is large enough. However, we do not make this approximation as empirical distributions of mutation effects are consistent with very low in different organisms (Martin and Lenormand 2006b). We then rewrite shom as a function of ι < 0 and y,
| (22) |
The bivariate {shom, ι} is then found by transforming the bivariate {y, ι} using (22) (combining the negative Gamma distribution of ι and the distribution of y above). The bivariate density Φ of {shom, ι} is defined in the range and is
| (23) |
where and are the parameters of the Gamma distribution of ι and K is a constant
| (24) |
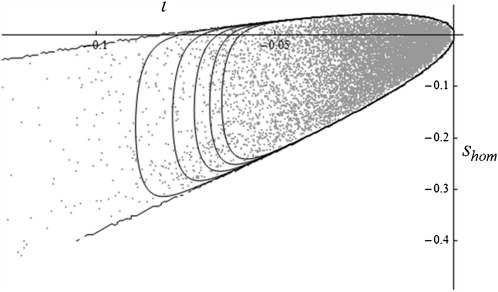
This distribution is illustrated on Figure 8. It could be used to provide a more powerful test of the present theory with bivariate data. Note that from Equation 23 the distribution of dominance can be derived among any subset of mutations, beneficial, deleterious, etc.
Figure 8.
Bivariate distribution Φ{ι,shom}. Illustration of the bivariate distribution Φ{ι, shom}, given in Equation 23 (black lines) with exact simulation of 10,000 mutants (gray dots). Parameter values are so = 0.04 and E(shom) = 0.05, ne = 3.5.
Empirical Test
Data survey
To test our theory, we surveyed the different estimates of average dominance that have been published in the last 40 years. The different data sets in our survey are listed in Table 1. We focused on the regression estimate of average dominance (hR in our notation), which was the most widely available. These data typically exclude large effect mutations. Thus, this estimator is also likely to be less biased compared to hM (see above). For instance, in Drosophila experiments (representing a large fraction of the available data), estimates are based only on the so called “quasi-normal” mutants. In our survey, we excluded two studies. We excluded the Ohnishi data (Ohnishi 1977a,b,c) because these experiments, although historically important, have been seriously questioned and because inconsistent dominance estimates were made depending on the method (Garcia-Dorado and Caballero 2000). We also excluded the Mackay et al. experiment (Mackay et al. 1992), because the results are inconsistent with basic observation expected in a mutation accumulation experiment: in this experiment the mutant lines do not exhibit more variance than the control lines after mutation accumulation. Other detailed information on each study is given in the legend of Table 1. Several other estimates of average dominance (range 0.15–0.35), based on segregating deleterious alleles in natural populations of flies and plants, have also been obtained (Lynch and Walsh 1998). As these estimates make strong assumptions such as mutation–selection equilibrium, they are not detailed here. Importantly, these data seem to quantitatively agree with the more direct estimates that we surveyed. To average results over studies, taking into account the standard error of each estimate and possible true variation between studies in average dominance, we used a random effect weighted mean meta-analysis (Gurevitch and Hedges 1999). We first averaged measures per studies and then across studies.
Table 1. Estimates of average dominance.
| hR | hM | Trait | Method Note | Analyzed | Species | Reference |
|---|---|---|---|---|---|---|
| 0.029 | — | Viability | MAa | Excluded | Drosophila melanogaster | Garcia-Dorado and Caballero (2000) |
| 0.10 | — | |||||
| 0.2 | — | |||||
| 0.328 | — | Viability | MA | Included | Chavarrias et al. (2001) | |
| −0.03 | — | Early ♀ fec. | MAb | Included | Houle et al. (1997) | |
| 0.12 | — | Late ♀ fec. | ||||
| 0.37 | — | ♂ longevity | ||||
| 0.26 | — | ♀ longevity | ||||
| −0.07 | — | ♂ mating ability | ||||
| 0.12 | — | Weighted mean | ||||
| 0.16 | 0.51 | Viability | TE | Included | Fry and Nuzhdin (2003) | |
| 0.01 | 0.45 | Viability | P-elem. | Excluded | Mackay et al. (1992) | |
| 0.21 | — | Viability | MAc | Included | Simmons and Crow (1977) | |
| 0.29 | — | Viability | MAc | Included | ||
| 0.40 | — | Viability | MAc | Included | ||
| 0.49 | — | Viability | MAc | Included | ||
| 0.21 | — | Viability | MAc | Included | ||
| 0.21 | — | Viability | MAc | Included | ||
| 0.40 | — | Viability | MAc | Included | ||
| 0.133 | 0.195 | Growth rate | SM | Included | Saccharomyces cerevisiae | Szafraniec et al. (2003) |
| 0.64 | — | Productivity | MA | Included | Caenorhabditis elegans | Vassilieva et al. (2000) |
| 0.05 | — | Surviv. to mat. | ||||
| −0.10 | — | Longevity | ||||
| 0.55 | — | Intrinsic rate incr. | ||||
| 0.48 | — | Convergence rate | ||||
| 0.69 | — | Generation rate | ||||
| −0.508 | 0.12 | Productivity | EMSd | Included | Peters et al. (2003) | |
| −0.508 | 0.08 | Relative fitness |
hM is given by E(shet)/E(shom) and hR by Cov(shet,shom)/Var(shom), where shet and shom are the distributions of the heterozygous and homozygous fitness effects of mutations, respectively. The MA method refers to mutation accumulation of experiments that produce lines with unknown numbers of spontaneous mutations. EMS represents the same method but with mutagen-induced mutations. TE represents transposable elements. P-elem represents for P elements and SM represents the study of lines carrying exactly one single mutation. Different proxy for fitness are used in the experiments (viability, productivity, growth rate, indicated in the Trait column).
Although historically important, Ohnishi estimates present several problems as explained in Garcia-Dorado and Caballero (2000). Originally, hR values were estimated to be low and inconsistent between crossing schemes (0.1 in coupling crosses and 0.029 in repulsion crosses). The reanalysis in Garcia-Dorado and Caballero (2000) of the same data yields a much higher value (0.2). Given all these uncertainties and the fact that other Drosophila estimates are available, we excluded this data set from our analysis.
The weighted mean (0.12) given in the table is simply an average over traits given in Houle et al. (1997). We did not include it in our composite estimate of average dominance.
Simmons and Crow (1977) report recalculated values of hR for different Mukai’s experiments.
In this experiment, overdominance was observed in several sublines. The overall pattern [Figure 3 in Peters et al (2003)] was strikingly similar to what our model would predict away from the optimum (hR ≈ 0.5 and presence of overdominance, Figure 5, right) although hM was lower (≈ 0.1). Note that the hR estimate given in the article (0.02) is mistaken (A. Peters, personal communication).
Quantitative agreement to the theory
As mentioned above, hR is computed over a set of mutations as the slope of the regression of shet on shom. Here, shom = log(whom) and shet = log(whet) denote the log-fitness relative to that of the nonmutated initial genotype of a homozygous and heterozygous mutant, respectively. Again, our model predicts
| (13) |
where, again, ε = so/|E(shom)| and measures the level of adaptation of the wild type, in the environment considered. More precisely, it is the fitness distance between the optimum and the wild type (so), scaled to the absolute value of average homozygous effect of mutations |E(shom)|. At the optimum (ε = 0), hR equals one-quarter in the additive model (υ = 1/2) and tends toward one-third in the extreme situation where υ is uniformly distributed in the range [0, 1]. As the distance to the optimum becomes larger, hR tends toward one-half. Overall, our model predicts that hR should be in the range [1/4, 1/2] and closer to 1/4 when measured under optimal conditions (such as laboratory conditions on model organisms).
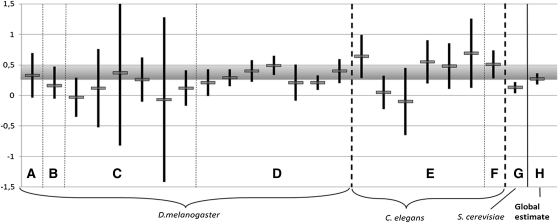
Figure 9 illustrates the different estimates as well as their confidence interval, which agrees with our theory and does not point toward strong departure from strict additivity in the model. In all experiments except one, the average measure of dominance does not significantly differ from our prediction (Figure 9). However, due to the large uncertainty in the estimates from most studies, a meta-analysis provided a more powerful evaluation of the prediction. We found that across studies, average dominance was hR = 0.27 (confidence interval [0.18, 0.36]). Other qualitative surveys (Charlesworth and Charlesworth 1998; Szafraniec et al. 2003) also gave one-quarter as an average measure of dominance of mildly deleterious mutations. The estimates of average dominance based on segregating deleterious alleles in natural populations of flies and plants (range 0.15–0.35) were not included in this survey but are also in agreement with this figure (Lynch and Walsh 1998). Computing this average dominance does not contradict the fact that average dominance varies to some extent among studies (although much of the variation arises due to within-studies measurement error). Additional and more precise studies would help in quantifying this aspect. Beyond the average measure of dominance, our model predicts that overdominant mutations can easily be observed when away from the optimum. This is precisely what Peters et al. (2003) found when studying the effect of mutations in Caenorhabditis elegans (Table 1). The power of such tests based on the average estimate of dominance is necessarily limited, and we believe that more insights could be gained by comparing whole distributions of homo- and heterozygous fitness effects (or shom and ι) with explicit predictions for this distribution (Equation 23, Figure 8). Such an approach would also avoid the problem of the different possible definition of dominance.
Figure 9.
Survey of empirical estimates of average dominance. We confronted our prediction (hR within [0.25–0.5] and closer to 0.25, shaded region) with different empirical estimates of hR (with their confidence interval), across species and different traits. D. melanogaster: (A) viability (Chavarrias et al. 2001), (B) viability (Fry and Nuzhdin 2003), (C) female early fecundity, female late fecundity, male longevity, female longevity, male mating ability, weighted mean (not used to calculate our composite estimate of average dominance) (Houle et al. 1997), (D) viability, recalculated hR from different Mukai’s experiments (Simmons and Crow 1977). C. elegans: (E) productivity, survival to maturity, longevity, intrinsic rate of increase, convergence rate, generation rate (Vassilieva et al. 2000), (F) relative fitness (Peters et al. 2003). Saccharomyces cerevisiae, (G) growth rate (Szafraniec et al. 2003). (H) Composite weighted estimate of hR across studies 0.27 [CI 0.18–0.36].
Mutations of large effect
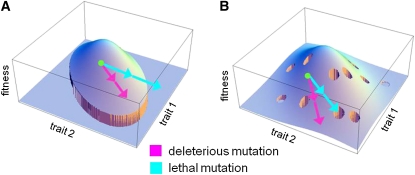
One limit of the simplified landscape model proposed so far is that, because it focuses on log-fitness on a smooth landscape, it naturally ignores “ruggedness” and abrupt fitness effects. In particular, this approach ignores lethal or large-effect mutations despite the fact that they represent a large fraction of newly arising mutations (Fudala and Korona 2009). This may not be problematic since most of these mutations may be also excluded from mutation accumulation experiments that we have surveyed. However, mutations of large effect do occur and it is interesting to see whether their fitness effect can also be understood in a landscape framework. There are two extreme ways to introduce this class of mutations. The first, maybe most intuitive, is to consider that the fitness function is truncated for some extreme trait values (Figure 10A), and the second is to consider that lethal mutations are caused by random genetic incompatibilities unrelated to the trait values as depicted in “holey landscapes,” (Gavrilets 1997) (Figure 10B). Note that we depict this situation with “holes” for the ease of illustration; it does not mean that some specific combinations of trait values have a low fitness. It means only that genetic incompatibilities may arise for some mutations in some backgrounds, somewhat independent of trait values. In the first case, the heterozygous effect of strongly deleterious mutations should always be greater than that of mildly deleterious ones. Indeed mutations that are far enough from the optimum to be strongly deleterious in two copies must be already far from the optimum in one copy (hence have higher than average heterozygous effect). In the second case, because holes are independent of trait effects, heterozygous effects of strongly and mildly deleterious mutations should be drawn from the same distribution and thus have the same mean value. The data on lethal mutations in Drosophila (Simmons and Crow 1977) and yeast (Szafraniec et al. 2003) clearly support the second possibility. The data on the fitness effect of gene deletion in yeast (Steinmetz et al. 2002) also show that strongly deleterious deletion (around 20% of the deletions) have independent heterozygous and homozygous effect as indicated by the analysis of Agrawal and Whitlock (2011). Using their “model7” (providing the best fit to the data), mutations with s = 0.05 have hs = 0.013 while mutations with s = 0.5 (a 10-times increase) have hs = 0.014 (a 1.06 times increase). In other words, mean heterozygote fitness is constant for a wide variation of s values within the minority of deletion of large effect. Such a flat relationship is exactly what is predicted if the ruggedness on homozygotes takes the form of holes as we intuitively present it (Figure 10B).
Figure 10.
Alternative models for mutations of large effect and lethals. (A) The fitness function is truncated for extreme trait values: any mutation with effect larger than some fitness threshold has a much stronger deleterious effect than with a smooth landscape model. As a consequence, the average heterozygous effect of mutations of weak homozygous effect (pink) is necessarily smaller than that of mutations of large homozygous (blue). (B) Mutations of large effects and lethals result from genetic incompatibilities unrelated to the trait values. For the sake of illustration, these incompatibilities are illustrated as small random holes on the fitness surface, but these holes are not necessarily a fixed feature of the fitness surface, they may differ across genetic background or environments. In this case the average heterozygous effect of mild and strongly deleterious mutations (including lethal) is equal on average.
These observations show first that a smooth landscape model is not sufficient to account for the distribution of mutations of large effects. Second they show that incorporating ruggedness in the form of holes (rather than thresholds) may account for the distribution of effect of this class of mutations. This is not so surprising; genetic incompatibilities are commonplace as revealed by recent progress on the genetics of speciation (Orr and Presgraves 2000) or the observation that species can often have very similar phenotypes and yet cannot interbreed. Of course, this does not exclude the possibility that extreme trait values could be lethal or that environmental variation can cause some form of lethal fitness threshold too (Figure 10a), but it does suggest that incompatibilities are the main source of strongly deleterious mutation in the laboratory experiments mentioned above. New data would be valuable to further study the difference in behavior between mutation of small and large effect (including lethals).
Discussion
We propose a theory for the dominance of mutations rooted in the idea of a smooth and relatively simple fitness landscape. This approach is clearly a compromise between precision, realism, and generality (Levins 1966). Such a balance is well suited to evolutionary genetics, but may be less appropriate in different circumstances. Before trying to present the advantages and main statements of this theory in particular relative to previous ones, we can try to make clear what the limits of such a theory are. First, this approach applies to the effect of the bulk of newly arising mutations of moderate phenotypic effects. It does not really address the dominance of alleles maintained highly polymorphic at particular loci. Second, as we have seen, it does not directly incorporate, but can accommodate, the idea of ruggedness, which might be essential to understanding the properties of mutation of strong effect and lethals. We return to these issues below. On the other hand, the central advantages of this theory are that (i) it provides a testable, quantitative prediction that seems to match the available data on average dominance of mildly deleterious mutations with an economy of assumptions; and (ii) it extends well beyond the question of dominance and accounts for a variety of mutation effects. We discuss these issues below. We also discuss what our theory brings to recurrent question/controversies about dominance, such as the h–s correlation, the phenomenon of overdominance, and the dominance of fitness vs. nonfitness traits.
A quantitative agreement
Contrary to previous theories, our theory makes a quantitative and testable prediction about the average dominance of mildly deleterious mutations, which is in good agreement with the available data (Figure 9). However, these predictions are open to further tests, in particular regarding the overall distribution of dominance (Equation 23 and Figure 8), the effect of the environment (distance to the optimum so; see Equation 13 and Figure 5) or the properties of lethal and large-effect mutations. This quantitative agreement is important for several reasons. The debate over dominance involved long discussions, argument, and counterarguments. However, despite the long-lasting history of the controversy, there is no quantitative theory for dominance to which we could confront the present one. Fisher’s, Wright’s, and metabolic theories and all their subsequent developments do not provide a prediction for the distribution of dominance; they state only that it should be less than one-half. This is not to say that these theories do not point toward relevant and interesting biological phenomena, but that they remain qualitative and therefore more difficult to directly confront with data. We hope that a merit of the present approach is to offer a clearly testable alternative in a debate that we feel has remained too qualitative over the last 80 years. It is, however, important to note that we only crudely compared our predictions to data (i.e., we only surveyed measures of average dominance, and among the subset of mildly deleterious mutations). The present theory could be tested more extensively, and with more power of rejection, provided additional detailed data become available on the full distribution of dominance of mutations on fitness.
Phenotype to fitness map
This theory does not focus exclusively on enzymes so that it appears to make less strong assumptions on the phenotype–fitness map than metabolic theories do. The assumptions made in landscape models, on the other hand, are often considered less intuitive or less “biological” than metabolic descriptions. The model does assume a given phenotype–fitness map, but using the general concept of stabilizing selection on a small phenotypic local scale (hence a treatment of weak effect mutations here). We believe this provides generality compared to making assumptions about the adaptive value of specific (metabolic) traits, especially a linear relationship between metabolic flux and fitness (see Introduction). Fitness integrates processes occurring well beyond metabolism (development, morphology, resource acquisition and allocation, mate choice, and so on) and we believe that forcing all mutation effects into a metabolic context limits the generality of the conclusions, even if an important part of fitness affecting mutations are probably related to metabolism. Like Fisher’s theory, this model requires natural selection, but in a different way: natural selection ensures that most of the time and for most of their traits, populations will not be too far from their phenotypic optimum, so that the fitness surface is likely to be, at least locally, concave and quadratic in first approximation. The concavity of fitness surfaces, which causes recessivity in our model, is further supported by the fact that mutations are always deleterious on average (Martin and Lenormand 2006a). However, like metabolic theory, this model avoids Fisher’s assumption that the dominance of each wild-type allele is molded by natural selection. In any case, as Mayo and Burger advocated (Mayo and Burger 1997), a general theory for the dominance of newly arising mutations does not preclude that dominance could evolve in specific circumstances at selected loci exhibiting high polymorphism (see review in Bagheri and Wagner 2004 and Llaurens et al. 2009; Schoen and Busch 2009; Chang and Noor 2010). Dominance can evolve perfectly in some cases, but this may not cause the bulk of newly arising mutations to be recessive.
h–s correlation
It has been shown for a long time and in several experiments that lethal mutations were much more recessive than mutations of small effects (Simmons and Crow 1977). This observation led to the idea that there is a widespread h–s correlation. Because such a relationship is not expected in Fisher’s theory, it was used against it, as we already explained (Charlesworth 1979). On the contrary, in the metabolic theory, such a relationship holds for integrated metabolic traits (such as flux or intermediate substrate concentration). Thus, metabolic theory is qualitatively compatible with the observed h–s correlation. More recent analyses on the yeast deletion database (Steinmetz et al. 2002) have also shown the presence of a widespread h–s correlation. However, the same data have been interpreted in favor either of the metabolic theory (Phadnis and Fry 2005) or of models that depend on indirect selection on homeostatic gene expression (Agrawal and Whitlock 2011). At this point, it is important to underline that the evidence for a h–s correlation relies on mutations of large effects (say with s above 0.05–0.1), which represent a minority of all mutations, and it is still unclear that there is a h–s correlation within mutations of small effects. For instance in the yeast deletion database mentioned above, and as far as these data can be trusted (see below), 70–80% of the mutations have small effects and do not exhibit a h–s correlation and the prediction derived from a smooth landscape model would apply to these mutations. For the remaining mutations it is certainly simpler to say that heterozygous and homozygous effects are independent than to say that there is a h–s correlation [see above the analysis of Agrawal and Whitlock (2011) of the yeast deletion data]. In this regard, it can be noted that while metabolic theory predicts h–s correlations, it cannot easily account for independent homo- and heterozygotes effects.
As we have shown, a strong h–s correlation is not expected in a smooth landscape model, especially close to an optimum (Equation 13), and measuring such correlation among mild mutations could be an interesting way to evaluate this prediction. However, adding some ruggedness in such models generates this correlation (see above and Figure 10) among mutations of large effect. Overall, it remains quite plausible that different phenomena arise for mutations of large and small effects. The high-throughput data measured on yeast gene deletion collection (Steinmetz et al. 2002) would have been a good starting point to investigate this issue quantitatively as in Phadnis and Fry (2005) and Agrawal and Whitlock (2011). However, this data set presents internal inconsistencies that preclude, in our opinion, using them quantitatively, especially for the bulk of mutation of small effects that are relevant to testing the smooth landscape theory presented here. Repeatability is very poor among mutations of small effects and unexpectedly lower between replicates in the same environment than across environments (sometimes with negative correlations among replicate measurements). The conclusions drawn in previous studies of this data set (Phadnis and Fry 2005; Agrawal and Whitlock 2011) may hold in spite of these problems, because they dealt mainly with the subset of strongly deleterious effects (∼20% of deletions) where repeatability is better. Clearly, more work is needed to better understand the dominance of mutations of large effects. The theory we outline in Figure 10B seems to qualitatively agree with the available evidence and it may help generate simple quantitative predictions. It may also serve to provide a framework with which to further model genetic incompatibilities and underdominance (Barton 2001) or to further investigate the variation of dominance across broad functional categories of genes (Kondrashov and Koonin 2004; Phadnis and Fry 2005; Agrawal and Whitlock 2011).
Overdominance
One limit of previous theories is that they do not provide any simple mechanism allowing for overdominance (see, e.g., Omholt et al. 2000 for metabolic theory). This theory does provide such mechanism. To the extent that stabilizing selection occurs on a trait, overdominance is indeed almost inevitable as long recognized by Fisher, Wright, Haldane, and Muller (Crow 1987). Fitness landscape theory predicts that overdominance could be observed when studying more particularly the effect of new mutations on fitness. Fitness overdominance is observed (Simmons and Crow 1977; Mitchell-Olds 1995; Peters et al. 2003; Zeyl et al. 2003) when studying the effects of new mutations, even if its importance is often minimized for technical, methodological (Fry 2004), or historical reasons. On the other hand, the literature on the maintenance of variation has largely argued against a predominant role of overdominance (Crow 1987). However, the existence of overdominance among newly arising mutations does not preclude that they play a minimal role in the maintenance of variation. With the stabilizing selection model assumed here, overdominance depends on the genetic background so that it would hardly maintain polymorphism at more than one locus per independently selected trait if these backgrounds vary, i.e., if there is genetic variation at other loci (Hastings and Hom 1989). In addition, even if a single locus segregates, a gene duplication can rapidly fix the heterotic gene combinations, which would suppress the heterozygous advantage (Haldane 1932). Thus, distinguishing overdominance in newly arising mutations from overdominance in standing variation is probably an important step toward clarifying this debate. Furthermore, the present landscape theory, by dealing with both deleterious recessive mutations and overdominant mutations, may help bridge the gap between the Fisher–Wright debate over dominance and the Muller–Dobzhansky debate over overdominance (Crow 1987) that have developed in parallel despite addressing related issues.
Fitness vs. traits
The fitness landscape approach helps clarify the scope of a possible theory of dominance, which, in its current form, can be related only to fitness. This does not imply that we cannot get theoretical insights into the dominance of some particular characters, such as metabolic flux, as has been successfully done by metabolic theories. However, we do believe that (i) many issues related to dominance in evolutionary genetics depend on dominance for fitness, not for any other character, and (ii) it is difficult to provide general predictions for arbitrary characters that are not related to fitness. Indeed, strictly speaking, the distribution of dominance of characters in general is not something that can be defined unambiguously. This point caused considerable confusion, so that it is necessary to state it very clearly. The definition of characters is somewhat arbitrary. One can study size or weight of individuals in a population, but equally valid is the study of log(size), log(weight), or whatever function of size and weight one wishes. The set of arbitrarily defined traits is infinite and the dominance of “traits” will depend on these alternative definitions. If an allele changes additively a particular trait x, it will exhibit a possibly entirely different dominance when applied to f(x), where f stands for any function. This does not say that it is not perfectly valid and meaningful to compute the dominance of an allele with respect to a particular well-defined trait. It says only that we cannot aggregate measures of dominance on different characters without a criterium on the nature of the character.
Since the definition and measuring scale of characters are arbitrary, the distribution of dominance is meaningful only if it is sampled across mutations for the same character and not across characters. This ambiguity has plagued the debate on dominance on several occasions. For instance, in his first article on dominance, Fisher (see the data compiled in Fisher 1928) argues on the recessivity of mutations by compiling a data set on dominance on different characters not related to fitness, which is not relevant to his own theory. Wright made an important step by implicitly considering the dominance on a secondary character depending on the dominance of a primary character, where the secondary character is defined as a squared deviation of the primary character from an optimum (Wright 1935). However, this approach was unrelated to his theory of dominance and did not lead him to revise his metabolic view and more generally the view that a theory of dominance for arbitrary traits could be achieved. Similarly, it is unclear why metabolic theory could apply to character in general and fitness in particular. It applies only to a limited set of traits (flux, etc.) that are not typically the ones measured when average dominance of mutation is investigated. For instance, even within the theory, different characters exhibit different dominance (enzyme activity is additive, whereas flux, substrate concentrations, and so on are recessive).
Another example is the study by Orr (1991) who compiled the dominance of mutations on different characters in the haploid Chlamydomonas. This compilation led to falsification of Fisher’s theory and was quite influential in this respect. One important limit of this study was that the traits chosen, although several were probably under selection, were not chosen to be proxies for fitness. As a last example, we counted the number of single-locus genetic disorders described as being recessive or dominant in humans (OMIMdatabase, http://www.ncbi.nlm.nih.gov/sites/entrez?db+omim). In this database, a roughly equal proportion of mutations are classified as being dominant and recessive, which seems entirely different from the usual finding in other species (see our survey). However, it is unlikely that deleterious mutations in humans exhibit different dominance. A likely explanation for this discrepancy is rather that the definition of symptoms varies from case to case and differ from the fitness effect. A disease typically occurring late in life has little impact on fitness, for example, and more generally symptom gravity may be poorly correlated with fitness effect for many illnesses (see a similar argument in Motulsky 2010). In our theory, the mathematical argument can hold for any trait (not only fitness) that can be thought of as the optimization of a set of underlying primary traits. However, only a measure of fitness can be thought as being comparable for different organisms in different environments. This is not contradicted by the fact that measuring fitness is often imperfect as measured by a combination of life-history traits. This is only the best of what can be done.
Generality
Finally, the present landscape theory is not tailored to explaining dominance. It also applies more generally to describing other aspects of mutational effects; for example, it provides empirically valid predictions for epistasis between mutations (Martin et al. 2007), the variation in mutational effects across environments (Martin and Lenormand 2006a), and the shape of the distribution of mutational effects (Martin and Lenormand 2006b). Fitness landscape models, whether one considers them oversimplifying or not, at least offer a general, robust, and importantly testable framework within which to understand the fitness effect of mutations of small effects. There are different strategies for model building (Levins 1966; Orzack and Sober 1993), and the top-down approach presented here will certainly enrich the debate between the polarized views of metabolic and selective theories.
Acknowledgments
We thank S. Billiard, D. Bourguet, L.-M. Chevin, G. Epinat, M. Kirkpatrick, R. Korona, S. Otto, A. Peters, O. Ronce, C. Sharfe, L. Steinmetz, and two anonymous reviewers for comments and discussion. This work was supported by European Research Council starting grant ‘QuantEvol’ to T.L.
Literature cited
- Agrawal A. F., 2009. Spatial heterogeneity and the evolution of sex in diploids. Am. Nat. 174: S54–S70 [DOI] [PubMed] [Google Scholar]
- Agrawal A. F., Whitlock M. C., 2011. Inferences about the distribution of dominance drawn from yeast gene knockout data. Genetics 187: 553–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagheri H., Wagner G., 2004. Evolution of dominance in metabolic pathways. Genetics 168: 1713–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton N. H., 2001. The role of hybridization in evolution. Mol. Ecol. 10: 551–568 [DOI] [PubMed] [Google Scholar]
- Bataillon T., Kirkpatrick M., 2000. Inbreeding depression due to mildly deleterious mutations in finite populations: size does matter. Genet. Res. 75: 75–81 [DOI] [PubMed] [Google Scholar]
- Bourguet D., 1999. The evolution of dominance. Heredity 83: 1–4 [DOI] [PubMed] [Google Scholar]
- Bourguet D., Prout M., Raymond M., 1996. Dominance of insecticide resistance presents a plastic response. Genetics 143: 407–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürger R., Bagheri H., 2008. Dominance and its evolution, pp. 945–952 Encyclopedia of Ecology, edited by Se J., Bd F. Elsevier, Oxford [Google Scholar]
- Cailleau A., Cheptou P.-O., Lenormand T., 2010. Ploidy and the evolution of endosperm of flowering plants. Genetics 184: 439–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A. S., Noor M. A. F., 2010. Epistasis modifies the dominance of loci causing hybrid male sterility in the Drosophila pseudoobscura species group. Evolution 64: 253–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., 1979. Evidence against Fisher’s theory of dominance. Nature 278: 848–849 [Google Scholar]
- Charlesworth B., Charlesworth D., 1998. Some evolutionary consequences of deleterious mutations. Genetica 103: 3–19 [PubMed] [Google Scholar]
- Charlesworth D., Willis J. H., 2009. Fundamental concepts in genetics: the genetics of inbreeding depression. Nat. Rev. Genet. 10: 783–796 [DOI] [PubMed] [Google Scholar]
- Chavarrias D., Lopez-Fanjul C., Garcia-Dorado A., 2001. The rate of mutation and the homozygous and heterozygous mutational effects for competitive viability: a long-term experiment with Drosophila melanogaster. Genetics 158: 681–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevin L. M., Martin G., Lenormand T., 2010. Fisher’s model and the genomics of adaptation: restricted pleiotropy, heterogeneous mutation and parallel evolution. Evolution 64: 3213–3231 [DOI] [PubMed] [Google Scholar]
- Crow J., 1987. Muller, Dobzhansky, and overdominance. J. Hist. Biol. 20: 351–380 [Google Scholar]
- de Visser J., Hermisson J., Wagner G. P., Meyers L. A., Bagheri H. C., et al. , 2003. Perspective: evolution and detection of genetic robustness. Evolution 57: 1959–1972 [DOI] [PubMed] [Google Scholar]
- Dykhuizen D. E., Dean A. M., Hartl D. L., 1987. Metabolic flux and fitness. Genetics 115: 25–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epinat G., Lenormand T., 2009. The evolution of assortative mating and selfing with in- and outbreeding depression. Evolution 63: 2047–2060 [DOI] [PubMed] [Google Scholar]
- Fisher R. A., 1928. The possible modification of the response of the wild type to recurrent mutations. Am. Nat. 62: 115–126 [Google Scholar]
- Fry D. J., Nuzhdin V. S., 2003. Dominance of mutations affecting viability in Drosophila melanogaster. Genetics 163: 1357–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry J. D., 2004. How common are overdominant mutations? Genetics 167: 1031–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fudala A., Korona R., 2009. Low frequency of mutations with strongly deleterious but nonlethal fitness effects. Evolution 63: 2164–2171 [DOI] [PubMed] [Google Scholar]
- Garcia-Dorado A., Caballero A., 2000. On the average coefficient of dominance of deleterious spontaneous mutations. Genetics 155: 1991–2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilets S., 1997. Evolution and speciation on holey adaptive landscapes. Trends Ecol. Evol. 12: 307–312 [DOI] [PubMed] [Google Scholar]
- Gemmell N., Slate J., 2006. Heterozygote advantage for fecundity. PLoS ONE 1: e125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevitch J., Hedges L. V., 1999. Statistical issues in ecological meta-analyses. Ecology 80: 1142–1149 [Google Scholar]
- Haldane J. B. S., 1932. The Causes of Evolution. Harper, New York [Google Scholar]
- Hastings A., Hom C. L., 1989. Pleiotropic stabilizing selection limits the number of polymorphic loci to at most the number of characters. Genetics 122: 459–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houle D., Hughes K. A., Assimacopoulos S., Charlesworth B., 1997. The effects of spontaneous mutation on quantitative traits. II. Dominance of mutations with effects on life-history traits. Genet. Res. 70: 27–34 [DOI] [PubMed] [Google Scholar]
- Hurst L. D., Randerson J. P., 2000. Dosage, deletions and dominance: simple models of the evolution of gene expression. J. Theor. Biol. 205: 641–647 [DOI] [PubMed] [Google Scholar]
- Kacser H., Burns J. A., 1981. The molecular basis of dominance. Genetics 97: 639–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov F. A., Koonin E. V., 2004. A common framework for understanding the origin of genetic dominance and evolutionary fates of gene duplications. Trends Genet. 20: 287–291 [DOI] [PubMed] [Google Scholar]
- Lande R., 1976. Natural-selection and random genetic drift in phenotypic evolution. Evolution 30: 314–334 [DOI] [PubMed] [Google Scholar]
- Levins R., 1966. The strategy of model building in population biology. Am. Sci. 54: 421–431 [Google Scholar]
- Llaurens V., Billiard S., Castric V., Vekemans X., 2009. Evolution of dominance in sporophytic self-incompatibility systems. I. Genetic load and coevolution of levels of dominance in pollen and pistil. Evolution 63: 2427–2437 [DOI] [PubMed] [Google Scholar]
- Lynch M., Walsh B., 1998. Genetics and Analysis of Quantitative Traits. Sinauer, Sunderland [Google Scholar]
- Lynch M., Blanchard J., Houle D., Kibota T., Schultz S., et al. , 1999. Perspective: spontaneous deleterious mutation. Evolution 53: 645–663 [DOI] [PubMed] [Google Scholar]
- Mackay T. F. C., Lyman F. R., Jackson M. S., 1992. Effects of P elements insertions on quantitative traits in Drosophila melanogaster. Genetics 130: 315–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G., Lenormand T., 2006a The fitness effect of mutations across environments: a survey in the light of fitness landscape models. Evolution 60: 2413–2427 [PubMed] [Google Scholar]
- Martin G., Lenormand T., 2006b A general multivariate extension of Fisher's geometrical model and the distribution of mutation fitness effects across species. Evolution 60: 893–907 [PubMed] [Google Scholar]
- Martin G., Elena S. F., Lenormand T., 2007. Distributions of epistasis in microbes fit predictions from a fitness landscape model. Nat. Genet. 39: 555–560 [DOI] [PubMed] [Google Scholar]
- Mayo O., Burger R., 1997. The evolution of dominance: a theory whose time has passed? Biol. Rev. Camb. Philos. Soc. 72: 97–110 [Google Scholar]
- Mitchell-Olds T., 1995. Interval mapping of viability loci causing heterosis in Arabidopsis. Genetics 140: 1105–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motulsky A., 2010. Formal genetics of humans: mode of inheritance, pp. 165–210 Human Genetics, edited by Speicher M., Antonarakis S., Motulsky A. Springer, Berlin [Google Scholar]
- Ohnishi O., 1977a Spontaneous and ethyl methanesulfonate-induced mutations controlling viability in Drosophila melanogaster. 1. Recessive lethal mutations. Genetics 87: 519–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi O., 1977b Spontaneous and ethyl methanesulfonate-induced mutations controlling viability in Drosophila melanogaster. 2. Homozygous effect of polygenic mutations. Genetics 87: 529–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi O., 1977c Spontaneous and ethyl methanesulfonate-induced mutations controlling viability in Drosophila melanogaster. 3. Heterozygous effect of polygenic mutations. Genetics 87: 547–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omholt S., Plahte E., Oyehaug L., Xiang K., 2000. Gene regulatory networks generating the phenomena of additivity, dominance and epistasis. Genetics 155: 969–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr H. A., 1991. A test of fisher theory of dominance. Proc. Natl. Acad. Sci. USA 88: 11413–11415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr H. A., Otto S. P., 1994. Does diploidy increase the rate of adaptation? Genetics 136: 1475–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr H. A., Presgraves D. C., 2000. Speciation by postzygotic isolation: forces, genes and molecules. Bioessays 22: 1085–1094 [DOI] [PubMed] [Google Scholar]
- Orzack S. H., Sober E., 1993. A critical assessment of Levins’ “The Strategy of Model Building (1966)”. Q. Rev. Biol. 68: 533–546 [Google Scholar]
- Otto S. P., 2003. The advantages of segregation and the evolution of sex. Genetics 164: 1099–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto S. P., Bourguet D., 1999. Dominance in patchy environments. Am. Nat. 153: 561–574 [DOI] [PubMed] [Google Scholar]
- Otto S. P., Goldstein D. B., 1992. Recombination and the evolution of diploidy. Genetics 131: 745–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters A. D., Halligan D. L., Whitlock M. C., Keightley P. D., 2003. Dominance and overdominance of mildly deleterious induced mutations for fitness traits in Caenorhabditis elegans. Genetics 165: 589–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phadnis N., Fry J. D., 2005. Widespread correlations between dominance and homozygous effects of mutations: implications for theories of dominance. Genetics 171: 385–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provine W., 1986. Sewall Wright and Evolutionary Biology. University of Chicago Press, Chicago [Google Scholar]
- Roze D., Rousset F., 2005. Inbreeding depression and the evolution of dispersal rates: a multilocus model. Am. Nat. 166: 708–721 [DOI] [PubMed] [Google Scholar]
- Schoen D. J., Busch J. W., 2009. The evolution of dominance in sporophytic self-incompatibility systems. ii. Mate availability and recombination. Evolution 63: 2099–2113 [DOI] [PubMed] [Google Scholar]
- Simmons M. J., Crow J. F., 1977. Mutations affecting fitness in Drosophila populations. Annu. Rev. Genet. 11: 49–78 [DOI] [PubMed] [Google Scholar]
- Steinmetz L. M., Scharfe C., Deutschbauer A. M., Mokranjac D., Herman Z. S., et al. , 2002. Systematic screen for human disease genes in yeast. Nat. Genet. 31: 400–404 [DOI] [PubMed] [Google Scholar]
- Szafraniec K., Wloch D. M., Sliwa P., Borts R. H., Korona R., 2003. Small fitness effects and weak genetic interactions between deleterious mutations in heterozygous loci of the yeast Saccharomyces cerevisiae. Genet. Res. 82: 19–31 [DOI] [PubMed] [Google Scholar]
- Tenaillon O., Silander O. K., Uzan J. P., Chao L., 2007. Quantifying organismal complexity using a population genetic approach. PLoS ONE 2: e217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilieva L. L., Hook A. M., Lynch M., 2000. The fitness effects of spontaneous mutations in Caenorhabditis elegans. Evolution 54: 1234–1246 [DOI] [PubMed] [Google Scholar]
- Welch J. J., Waxman D., 2003. Modularity and the cost of complexity. Evolution 57: 1723–1734 [DOI] [PubMed] [Google Scholar]
- Whitlock M. C., 2002. Selection, load and inbreeding depression in a large metapopullation. Genetics 160: 1191–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S., 1929. Fisher’s theory of dominance. Am. Nat. 63: 274–279 [Google Scholar]
- Wright S., 1934. Physiological and evolutionary theories of dominance. Am. Nat. 67: 24–53 [Google Scholar]
- Wright S., 1935. The analysis of variance and the correlations between relatives with respect to deviations from an optimum. J. Genet. 30: 243–256 [Google Scholar]
- Zeyl C., Vanderford T., Carter M., 2003. An evolutionary advantage of haploidy in large yeast populations. Science 299: 555–558 [DOI] [PubMed] [Google Scholar]