Figure 2.
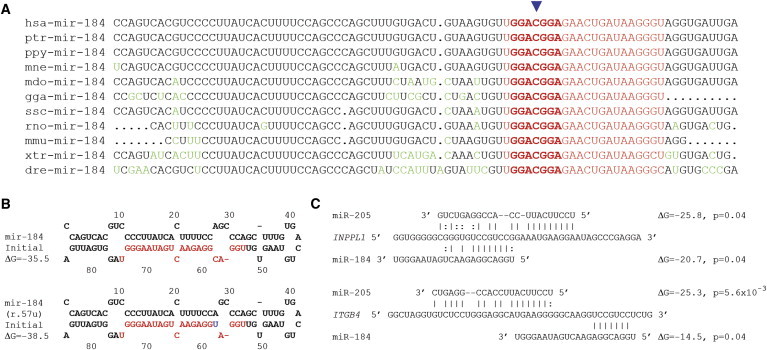
Variants of miR-184 and Conservation across Species, Structure of miR-184, miR-205 and Mutant miR-184, and Interactions of miR-184 and miR-205
(A) Multiple alignment of miR-184 sequences from different vertebrate species. The mutation site (miR-184 r.57c>u, indicated by the blue arrow) is fully conserved across all species. Changes from the human reference sequence are shown in green type; the mature miRNAs are shown in red type, and the seed regions are in bold. The following abbreviations are used: hsa, Homo sapiens; ptr, Pan troglodytes; ppy, Pongo pygmaeus; mne, Macaca nemestrina; mdo, Monodelphis domestica; gga, Gallus gallus; ssc, Sus scrofa; rno, Rattus norvegicus; mmu, Mus musculus; xtr, Xenopus tropicalis; dre, Danio rerio. Fifty-four additional species showed full conservation of the mature miR-184 sequence.
(B) Secondary structures and free energies of wild-type miR-184 and mutant miR-184 (r.57u) predicted by Mfold.
(C) Target sites of miR-184 and miR-205 in the 3′ UTRs of INPPL1 and ITGB4 predicted by MicroCosm with energies shown in kcal/mol.
