Figure 1.
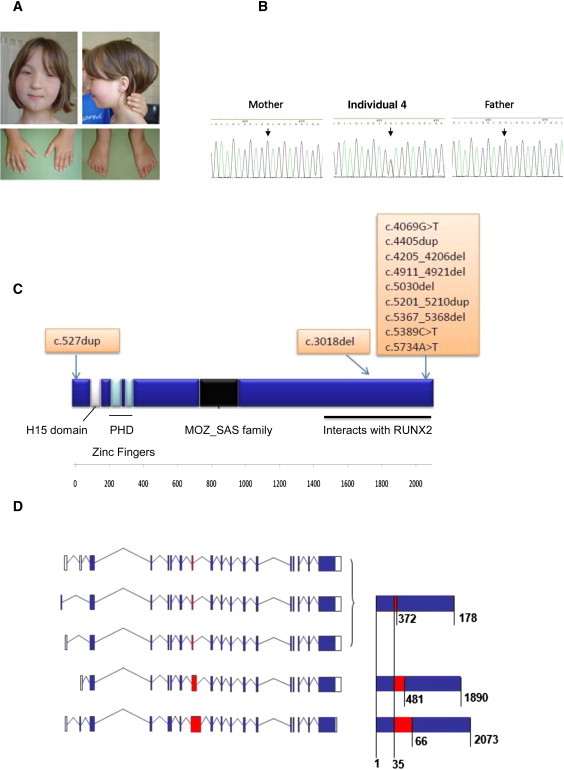
Clinical Phenotype, Structure, and Truncating Mutation of KAT6B
(A) Photos showing clinical features of individual 10 with typical SBBYSS. Facial features include severe blepharophimosis, a bulbous nasal tip, and a mask-like appearance. The thumbs and great toes are abnormally long and straight.
(B) Sanger sequencing of KAT6B in individual 4. The c.4069G>T (p.Glu1357X) mutation was absent in both parents and had arisen de novo.
(C) Schematic of KAT6B including the conserved domains and position of truncating mutations. The catalytic MYST domain of the MOZ-SAS family is the histoneacetyltransferase domain and contains a specific C2HC zinc finger together with an acetyl-coenzyme A binding domain. There is a conserved N-terminal domain ([NEMM] the N-terminal domain conserved in Enok) and a highly conserved C-terminal SM-rich domain. Mutations in typical SBBYSS individuals cluster in exon 18, the final exon, and leave the catalytic domain intact.
(D) KAT6B isoforms. Alternative splicing of KAT6B produces five protein coding transcripts. Three transcripts vary only in the 5′ UTR and produce the same mature protein. In-frame splicing within exon 8 results in three protein isoforms of 1781, 1890, and 2013 amino acids in length. As amino acid composition before and after exon 8 is identical in each isoform, the mutations found in exons 15 and 18 are assumed to have the same effect. White boxes indicate exons in UTR; blue boxes indicate translated exons; red box indicates exon 8; blue lines indicate introns. Mature protein contributed by exon 8 is shown in red; values indicate amino acid positions.
