Abstract
In the title molecule, C24H21ClN4OS2, the central 1,2,4-triazole ring forms dihedral angles of 89.05 (9), 86.66 (9) and 82.70 (10)° with the chloro-substituted benzene ring, the methylsulfanyl-substituted benzene ring and the phenyl ring, respectively. In the crystal, molecules are linked into sheets parallel to (100) by intermolecular N—H⋯N and weak C—H⋯O hydrogen bonds.
Related literature
For general background to and applications of 1,2,4-triazole derivatives, see: Holla et al. (2002 ▶, 2003 ▶); Rudnicka et al. (1986 ▶); Burch & Smith (1966 ▶); Kalyoncuoglu et al. (1992 ▶); Mhasalkar et al. (1970 ▶); Mir et al. (1970 ▶).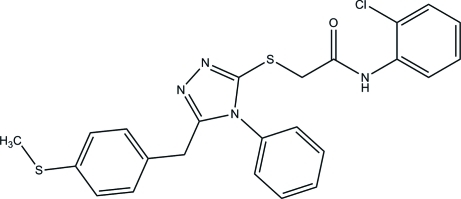
Experimental
Crystal data
C24H21ClN4OS2
M r = 481.02
Monoclinic,
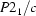
a = 14.2542 (7) Å
b = 16.3273 (9) Å
c = 10.1584 (6) Å
β = 96.372 (1)°
V = 2349.6 (2) Å3
Z = 4
Mo Kα radiation
μ = 0.37 mm−1
T = 297 K
0.52 × 0.27 × 0.22 mm
Data collection
Bruker APEXII DUO CCD diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2009 ▶) T min = 0.832, T max = 0.924
29877 measured reflections
7900 independent reflections
5235 reflections with I > 2σ(I)
R int = 0.028
Refinement
R[F 2 > 2σ(F 2)] = 0.049
wR(F 2) = 0.159
S = 1.05
7900 reflections
290 parameters
H-atom parameters constrained
Δρmax = 0.53 e Å−3
Δρmin = −0.45 e Å−3
Data collection: APEX2 (Bruker, 2009 ▶); cell refinement: SAINT (Bruker, 2009 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXTL (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXTL; molecular graphics: SHELXTL; software used to prepare material for publication: SHELXTL and PLATON (Spek, 2009 ▶).
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536811027565/lh5283sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536811027565/lh5283Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536811027565/lh5283Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1N1⋯N3i | 0.94 | 2.04 | 2.9787 (19) | 174 |
| C8—H8A⋯O1ii | 0.97 | 2.52 | 3.147 (2) | 123 |
| C11—H11B⋯O1iii | 0.97 | 2.47 | 3.416 (2) | 165 |
Symmetry codes: (i) 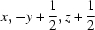 ; (ii)
; (ii) 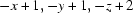 ; (iii)
; (iii) 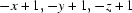 .
.
Acknowledgments
HKF and CSY thank Universiti Sains Malaysia for the Research University Grant 1001/PFIZIK/811160.
supplementary crystallographic information
Comment
1,2,4-Triazole compounds show a broad spectrum of biological activities, possibly due to the presence of the N–C–S group (Holla et al., 2002). Among the 1,2,4-triazoles, the mercapto-thione-substituted 1,2,4-triazole ring systems have been well studied and so far, a variety of biological activities have been reported for a large number of their derivatives, such as antibacterial (Burch & Smith, 1966), antifungal (Kalyoncuoglu et al., 1992), antitubercular (Mir et al., 1970), antimycobacterial (Rudnicka et al., 1986), anticancer (Holla et al., 2003) and hypoglycemic properties (Mhasalkar et al., 1970). In view of the above findings, and in continuation of our earlier work on the synthesis and biological activity of triazoles and their derivatives, we have synthesized a series of 1,2,4-triazole derivatives via joining a 1,2,4-triazole and acylamide group together in the same molecule and have studied their biological activities. As part of this research we have determined the crystal structure of th title compound.
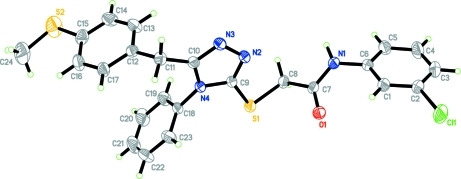
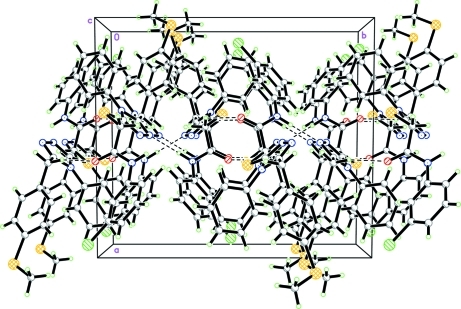
The molecular structure of the title compound (I) is shown in Fig. 1. The central 1,2,4-triazole ring forms dihedral angles of 89.05 (9), 86.66 (9) and 82.70 (10)° with the three benzene rings (C1–C6, C18–C23 and C12–C17). In the crystal, molecules are linked into double sheets parallel to (1 0 0) by intermolecular N1—H1N1···N3i, C8—H8A···O1ii and C11—H11B···O1iii hydrogen bonds (Table 1, Fig. 2).
Experimental
A equimolar mixture of 5-[4-(methylthiobenzyl]-4-phenyl-4H-[1,2,4]-triazole-3-thione (0.01 mol), 2-Chloro-N-(2-chlorophenyl)acetamide (0.01 mol) and dry potassium carbonate (0.01 mol) were refluxed for 6 h in 20 ml of absolute alcohol and excess of solvent was removed by distillation under reduced pressure. After cooling to room temperature, the reaction mixture was poured into 50 ml of water. The product which precipitated was filtered off, washed with methanol and dried. The crude product was re-crystallized from ethanol.
Refinement
The N-bound hydrogen atom was located from difference Fourier map and included in a riding-model approximation with N-H = 0.94Å and Uiso(H) = 1.2Ueq(N). All C-bound hydrogen atoms were positioned geomatrically [C–H = 0.93–0.97 Å] and refined using a riding model, with Uiso(H) = 1.2 or 1.5 Ueq(C). A rotating-group model were applied for methyl groups. Two reflections, (-1 1 1), and (6 6 4), were omitted.
Figures
Fig. 1.
The molecular structure of (I), with atom labels and 30% probability ellipsoids for non-H atoms.
Fig. 2.
The crystal packing of (I), showing the molecules linked into a double sheets parallel to (1 0 0). Hydrogen bonds are shown as dashed lines.
Crystal data
| C24H21ClN4OS2 | F(000) = 1000 |
| Mr = 481.02 | Dx = 1.360 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 7952 reflections |
| a = 14.2542 (7) Å | θ = 2.5–30.4° |
| b = 16.3273 (9) Å | µ = 0.37 mm−1 |
| c = 10.1584 (6) Å | T = 297 K |
| β = 96.372 (1)° | Block, brown |
| V = 2349.6 (2) Å3 | 0.52 × 0.27 × 0.22 mm |
| Z = 4 |
Data collection
| Bruker APEXII DUO CCD diffractometer | 7900 independent reflections |
| Radiation source: fine-focus sealed tube | 5235 reflections with I > 2σ(I) |
| graphite | Rint = 0.028 |
| φ and ω scans | θmax = 31.7°, θmin = 1.9° |
| Absorption correction: multi-scan (SADABS; Bruker, 2009) | h = −21→21 |
| Tmin = 0.832, Tmax = 0.924 | k = −24→23 |
| 29877 measured reflections | l = −14→14 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.049 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.159 | H-atom parameters constrained |
| S = 1.05 | w = 1/[σ2(Fo2) + (0.075P)2 + 0.4271P] where P = (Fo2 + 2Fc2)/3 |
| 7900 reflections | (Δ/σ)max < 0.001 |
| 290 parameters | Δρmax = 0.53 e Å−3 |
| 0 restraints | Δρmin = −0.45 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Cl1 | 0.90496 (5) | 0.48639 (5) | 1.17911 (9) | 0.1088 (3) | |
| S1 | 0.39474 (3) | 0.45413 (3) | 0.74551 (4) | 0.05346 (13) | |
| S2 | 0.00345 (5) | 0.24018 (6) | 0.06766 (9) | 0.1109 (3) | |
| O1 | 0.58819 (9) | 0.47636 (7) | 0.90982 (12) | 0.0551 (3) | |
| N1 | 0.59354 (9) | 0.34760 (8) | 0.99633 (14) | 0.0483 (3) | |
| H1N1 | 0.5589 | 0.2988 | 0.9997 | 0.058* | |
| N2 | 0.48488 (9) | 0.33585 (8) | 0.61649 (13) | 0.0469 (3) | |
| N3 | 0.48455 (9) | 0.30835 (8) | 0.48666 (14) | 0.0467 (3) | |
| N4 | 0.38196 (8) | 0.40807 (7) | 0.48700 (12) | 0.0393 (3) | |
| C1 | 0.74454 (12) | 0.40977 (11) | 1.08375 (18) | 0.0534 (4) | |
| H1A | 0.7464 | 0.4416 | 1.0081 | 0.064* | |
| C2 | 0.81439 (13) | 0.41592 (12) | 1.1889 (2) | 0.0623 (5) | |
| C3 | 0.81517 (16) | 0.36979 (15) | 1.3012 (2) | 0.0758 (6) | |
| H3A | 0.8632 | 0.3754 | 1.3704 | 0.091* | |
| C4 | 0.74302 (16) | 0.31485 (16) | 1.3089 (2) | 0.0766 (6) | |
| H4A | 0.7421 | 0.2828 | 1.3844 | 0.092* | |
| C5 | 0.67174 (13) | 0.30652 (12) | 1.20593 (19) | 0.0598 (4) | |
| H5A | 0.6239 | 0.2684 | 1.2119 | 0.072* | |
| C6 | 0.67148 (11) | 0.35481 (10) | 1.09415 (16) | 0.0449 (3) | |
| C7 | 0.55154 (11) | 0.41050 (10) | 0.92502 (14) | 0.0420 (3) | |
| C8 | 0.44971 (11) | 0.39121 (11) | 0.87723 (15) | 0.0469 (3) | |
| H8A | 0.4132 | 0.3957 | 0.9520 | 0.056* | |
| H8B | 0.4462 | 0.3346 | 0.8479 | 0.056* | |
| C9 | 0.42302 (10) | 0.39516 (9) | 0.61317 (15) | 0.0410 (3) | |
| C10 | 0.42372 (10) | 0.35210 (9) | 0.41135 (15) | 0.0411 (3) | |
| C11 | 0.40297 (11) | 0.34559 (11) | 0.26504 (15) | 0.0457 (3) | |
| H11A | 0.4454 | 0.3056 | 0.2332 | 0.055* | |
| H11B | 0.4161 | 0.3980 | 0.2260 | 0.055* | |
| C12 | 0.30248 (11) | 0.32128 (10) | 0.21711 (15) | 0.0429 (3) | |
| C13 | 0.26102 (13) | 0.25277 (12) | 0.2653 (2) | 0.0605 (5) | |
| H13A | 0.2949 | 0.2215 | 0.3308 | 0.073* | |
| C14 | 0.17027 (15) | 0.22984 (13) | 0.2183 (2) | 0.0679 (5) | |
| H14A | 0.1439 | 0.1834 | 0.2525 | 0.081* | |
| C15 | 0.11798 (13) | 0.27517 (13) | 0.1210 (2) | 0.0615 (5) | |
| C16 | 0.15828 (14) | 0.34441 (14) | 0.0742 (2) | 0.0656 (5) | |
| H16A | 0.1239 | 0.3765 | 0.0104 | 0.079* | |
| C17 | 0.24971 (13) | 0.36663 (12) | 0.12152 (17) | 0.0549 (4) | |
| H17A | 0.2760 | 0.4132 | 0.0879 | 0.066* | |
| C18 | 0.30298 (10) | 0.46012 (9) | 0.44437 (14) | 0.0411 (3) | |
| C19 | 0.21336 (12) | 0.43210 (13) | 0.4587 (2) | 0.0587 (4) | |
| H19A | 0.2049 | 0.3821 | 0.4996 | 0.070* | |
| C20 | 0.13671 (15) | 0.47902 (18) | 0.4118 (3) | 0.0819 (7) | |
| H20A | 0.0760 | 0.4605 | 0.4207 | 0.098* | |
| C21 | 0.14890 (19) | 0.55153 (18) | 0.3532 (3) | 0.0894 (8) | |
| H21A | 0.0965 | 0.5827 | 0.3219 | 0.107* | |
| C22 | 0.2370 (2) | 0.57963 (15) | 0.3394 (3) | 0.0892 (8) | |
| H22A | 0.2444 | 0.6300 | 0.2991 | 0.107* | |
| C23 | 0.31656 (15) | 0.53345 (12) | 0.3851 (2) | 0.0634 (5) | |
| H23A | 0.3770 | 0.5522 | 0.3753 | 0.076* | |
| C24 | −0.04042 (19) | 0.3111 (2) | −0.0555 (3) | 0.1131 (11) | |
| H24A | −0.1055 | 0.2992 | −0.0837 | 0.170* | |
| H24B | −0.0045 | 0.3070 | −0.1298 | 0.170* | |
| H24C | −0.0351 | 0.3655 | −0.0199 | 0.170* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cl1 | 0.0754 (4) | 0.0927 (5) | 0.1462 (7) | −0.0411 (3) | −0.0421 (4) | 0.0276 (4) |
| S1 | 0.0579 (2) | 0.0499 (2) | 0.0494 (2) | 0.01375 (18) | −0.00817 (17) | −0.00946 (17) |
| S2 | 0.0727 (4) | 0.1321 (7) | 0.1206 (6) | −0.0488 (4) | −0.0226 (4) | 0.0262 (5) |
| O1 | 0.0541 (7) | 0.0477 (6) | 0.0610 (7) | −0.0125 (5) | −0.0046 (5) | 0.0102 (5) |
| N1 | 0.0456 (7) | 0.0414 (7) | 0.0549 (8) | −0.0092 (5) | −0.0071 (6) | 0.0032 (6) |
| N2 | 0.0430 (6) | 0.0449 (7) | 0.0501 (7) | 0.0090 (5) | −0.0070 (5) | −0.0012 (5) |
| N3 | 0.0435 (6) | 0.0438 (7) | 0.0514 (7) | 0.0092 (5) | −0.0011 (5) | −0.0027 (5) |
| N4 | 0.0355 (5) | 0.0386 (6) | 0.0422 (6) | 0.0057 (5) | −0.0031 (4) | 0.0006 (5) |
| C1 | 0.0454 (8) | 0.0511 (10) | 0.0610 (10) | −0.0068 (7) | −0.0060 (7) | 0.0050 (8) |
| C2 | 0.0498 (9) | 0.0520 (10) | 0.0803 (13) | −0.0091 (8) | −0.0139 (8) | 0.0001 (9) |
| C3 | 0.0683 (12) | 0.0763 (14) | 0.0750 (14) | −0.0042 (11) | −0.0275 (10) | 0.0053 (11) |
| C4 | 0.0702 (13) | 0.0887 (16) | 0.0660 (12) | −0.0067 (12) | −0.0148 (10) | 0.0232 (11) |
| C5 | 0.0512 (9) | 0.0600 (11) | 0.0660 (11) | −0.0051 (8) | −0.0036 (8) | 0.0141 (9) |
| C6 | 0.0418 (7) | 0.0409 (8) | 0.0506 (8) | −0.0007 (6) | −0.0018 (6) | −0.0011 (6) |
| C7 | 0.0441 (7) | 0.0438 (8) | 0.0374 (7) | −0.0051 (6) | 0.0011 (5) | −0.0029 (6) |
| C8 | 0.0456 (8) | 0.0518 (9) | 0.0417 (7) | −0.0062 (7) | −0.0027 (6) | −0.0006 (6) |
| C9 | 0.0374 (6) | 0.0390 (7) | 0.0444 (7) | 0.0031 (5) | −0.0055 (5) | −0.0004 (6) |
| C10 | 0.0369 (6) | 0.0397 (7) | 0.0460 (7) | 0.0030 (5) | 0.0018 (5) | 0.0000 (6) |
| C11 | 0.0423 (7) | 0.0505 (9) | 0.0445 (8) | 0.0019 (6) | 0.0053 (6) | −0.0005 (6) |
| C12 | 0.0438 (7) | 0.0431 (8) | 0.0416 (7) | 0.0019 (6) | 0.0048 (6) | −0.0021 (6) |
| C13 | 0.0519 (9) | 0.0537 (10) | 0.0751 (12) | 0.0041 (8) | 0.0031 (8) | 0.0194 (9) |
| C14 | 0.0589 (10) | 0.0519 (11) | 0.0925 (15) | −0.0084 (9) | 0.0071 (10) | 0.0148 (10) |
| C15 | 0.0524 (9) | 0.0702 (12) | 0.0605 (10) | −0.0134 (9) | −0.0004 (8) | −0.0018 (9) |
| C16 | 0.0596 (10) | 0.0783 (13) | 0.0546 (10) | −0.0111 (9) | −0.0126 (8) | 0.0179 (9) |
| C17 | 0.0555 (9) | 0.0586 (10) | 0.0485 (9) | −0.0116 (8) | −0.0032 (7) | 0.0109 (7) |
| C18 | 0.0399 (7) | 0.0406 (7) | 0.0410 (7) | 0.0090 (6) | −0.0034 (5) | 0.0006 (6) |
| C19 | 0.0411 (8) | 0.0640 (11) | 0.0698 (11) | 0.0058 (8) | 0.0014 (7) | 0.0052 (9) |
| C20 | 0.0452 (10) | 0.1068 (19) | 0.0905 (16) | 0.0244 (11) | −0.0069 (10) | −0.0045 (14) |
| C21 | 0.0787 (16) | 0.0984 (19) | 0.0846 (16) | 0.0471 (14) | −0.0195 (12) | −0.0029 (14) |
| C22 | 0.120 (2) | 0.0542 (12) | 0.0880 (16) | 0.0259 (13) | −0.0135 (15) | 0.0193 (11) |
| C23 | 0.0667 (11) | 0.0490 (10) | 0.0731 (12) | 0.0034 (8) | 0.0010 (9) | 0.0127 (8) |
| C24 | 0.0639 (14) | 0.157 (3) | 0.111 (2) | −0.0165 (17) | −0.0253 (14) | 0.011 (2) |
Geometric parameters (Å, °)
| Cl1—C2 | 1.740 (2) | C10—C11 | 1.487 (2) |
| S1—C9 | 1.7371 (16) | C11—C12 | 1.514 (2) |
| S1—C8 | 1.7964 (16) | C11—H11A | 0.9700 |
| S2—C15 | 1.7574 (19) | C11—H11B | 0.9700 |
| S2—C24 | 1.767 (3) | C12—C17 | 1.377 (2) |
| O1—C7 | 1.2128 (19) | C12—C13 | 1.380 (2) |
| N1—C7 | 1.357 (2) | C13—C14 | 1.380 (3) |
| N1—C6 | 1.411 (2) | C13—H13A | 0.9300 |
| N1—H1N1 | 0.9408 | C14—C15 | 1.385 (3) |
| N2—C9 | 1.3075 (19) | C14—H14A | 0.9300 |
| N2—N3 | 1.3927 (19) | C15—C16 | 1.376 (3) |
| N3—C10 | 1.3038 (19) | C16—C17 | 1.386 (3) |
| N4—C9 | 1.3650 (18) | C16—H16A | 0.9300 |
| N4—C10 | 1.3717 (19) | C17—H17A | 0.9300 |
| N4—C18 | 1.4383 (17) | C18—C23 | 1.364 (2) |
| C1—C2 | 1.380 (2) | C18—C19 | 1.380 (2) |
| C1—C6 | 1.388 (2) | C19—C20 | 1.375 (3) |
| C1—H1A | 0.9300 | C19—H19A | 0.9300 |
| C2—C3 | 1.366 (3) | C20—C21 | 1.345 (4) |
| C3—C4 | 1.374 (3) | C20—H20A | 0.9300 |
| C3—H3A | 0.9300 | C21—C22 | 1.358 (4) |
| C4—C5 | 1.381 (3) | C21—H21A | 0.9300 |
| C4—H4A | 0.9300 | C22—C23 | 1.398 (3) |
| C5—C6 | 1.382 (2) | C22—H22A | 0.9300 |
| C5—H5A | 0.9300 | C23—H23A | 0.9300 |
| C7—C8 | 1.512 (2) | C24—H24A | 0.9600 |
| C8—H8A | 0.9700 | C24—H24B | 0.9600 |
| C8—H8B | 0.9700 | C24—H24C | 0.9600 |
| C9—S1—C8 | 98.04 (7) | C10—C11—H11B | 108.6 |
| C15—S2—C24 | 104.42 (12) | C12—C11—H11B | 108.6 |
| C7—N1—C6 | 125.38 (13) | H11A—C11—H11B | 107.6 |
| C7—N1—H1N1 | 117.3 | C17—C12—C13 | 117.67 (15) |
| C6—N1—H1N1 | 114.9 | C17—C12—C11 | 120.62 (15) |
| C9—N2—N3 | 106.43 (12) | C13—C12—C11 | 121.71 (15) |
| C10—N3—N2 | 108.13 (12) | C14—C13—C12 | 121.26 (17) |
| C9—N4—C10 | 104.81 (11) | C14—C13—H13A | 119.4 |
| C9—N4—C18 | 127.86 (12) | C12—C13—H13A | 119.4 |
| C10—N4—C18 | 126.78 (12) | C13—C14—C15 | 120.78 (18) |
| C2—C1—C6 | 118.11 (17) | C13—C14—H14A | 119.6 |
| C2—C1—H1A | 120.9 | C15—C14—H14A | 119.6 |
| C6—C1—H1A | 120.9 | C16—C15—C14 | 118.25 (17) |
| C3—C2—C1 | 122.97 (18) | C16—C15—S2 | 124.72 (16) |
| C3—C2—Cl1 | 118.34 (15) | C14—C15—S2 | 117.02 (15) |
| C1—C2—Cl1 | 118.68 (16) | C15—C16—C17 | 120.50 (18) |
| C2—C3—C4 | 118.09 (18) | C15—C16—H16A | 119.8 |
| C2—C3—H3A | 121.0 | C17—C16—H16A | 119.8 |
| C4—C3—H3A | 121.0 | C12—C17—C16 | 121.53 (17) |
| C3—C4—C5 | 120.9 (2) | C12—C17—H17A | 119.2 |
| C3—C4—H4A | 119.5 | C16—C17—H17A | 119.2 |
| C5—C4—H4A | 119.5 | C23—C18—C19 | 121.08 (16) |
| C4—C5—C6 | 120.03 (18) | C23—C18—N4 | 120.47 (15) |
| C4—C5—H5A | 120.0 | C19—C18—N4 | 118.37 (14) |
| C6—C5—H5A | 120.0 | C20—C19—C18 | 119.2 (2) |
| C5—C6—C1 | 119.86 (15) | C20—C19—H19A | 120.4 |
| C5—C6—N1 | 117.46 (15) | C18—C19—H19A | 120.4 |
| C1—C6—N1 | 122.64 (15) | C21—C20—C19 | 120.4 (2) |
| O1—C7—N1 | 124.62 (14) | C21—C20—H20A | 119.8 |
| O1—C7—C8 | 123.76 (15) | C19—C20—H20A | 119.8 |
| N1—C7—C8 | 111.39 (13) | C20—C21—C22 | 120.6 (2) |
| C7—C8—S1 | 116.35 (12) | C20—C21—H21A | 119.7 |
| C7—C8—H8A | 108.2 | C22—C21—H21A | 119.7 |
| S1—C8—H8A | 108.2 | C21—C22—C23 | 120.6 (2) |
| C7—C8—H8B | 108.2 | C21—C22—H22A | 119.7 |
| S1—C8—H8B | 108.2 | C23—C22—H22A | 119.7 |
| H8A—C8—H8B | 107.4 | C18—C23—C22 | 118.1 (2) |
| N2—C9—N4 | 110.87 (13) | C18—C23—H23A | 121.0 |
| N2—C9—S1 | 127.18 (11) | C22—C23—H23A | 121.0 |
| N4—C9—S1 | 121.93 (11) | S2—C24—H24A | 109.5 |
| N3—C10—N4 | 109.75 (13) | S2—C24—H24B | 109.5 |
| N3—C10—C11 | 126.37 (14) | H24A—C24—H24B | 109.5 |
| N4—C10—C11 | 123.86 (13) | S2—C24—H24C | 109.5 |
| C10—C11—C12 | 114.61 (13) | H24A—C24—H24C | 109.5 |
| C10—C11—H11A | 108.6 | H24B—C24—H24C | 109.5 |
| C12—C11—H11A | 108.6 | ||
| C9—N2—N3—C10 | 0.44 (17) | C9—N4—C10—C11 | −177.83 (14) |
| C6—C1—C2—C3 | 0.8 (3) | C18—N4—C10—C11 | 10.2 (2) |
| C6—C1—C2—Cl1 | −178.42 (14) | N3—C10—C11—C12 | 117.85 (17) |
| C1—C2—C3—C4 | 0.1 (4) | N4—C10—C11—C12 | −64.0 (2) |
| Cl1—C2—C3—C4 | 179.4 (2) | C10—C11—C12—C17 | 128.51 (17) |
| C2—C3—C4—C5 | 0.0 (4) | C10—C11—C12—C13 | −52.5 (2) |
| C3—C4—C5—C6 | −1.1 (4) | C17—C12—C13—C14 | 0.8 (3) |
| C4—C5—C6—C1 | 2.1 (3) | C11—C12—C13—C14 | −178.17 (18) |
| C4—C5—C6—N1 | −175.7 (2) | C12—C13—C14—C15 | 0.0 (3) |
| C2—C1—C6—C5 | −1.9 (3) | C13—C14—C15—C16 | −1.2 (3) |
| C2—C1—C6—N1 | 175.77 (17) | C13—C14—C15—S2 | 179.65 (18) |
| C7—N1—C6—C5 | 141.23 (18) | C24—S2—C15—C16 | 1.5 (3) |
| C7—N1—C6—C1 | −36.5 (3) | C24—S2—C15—C14 | −179.4 (2) |
| C6—N1—C7—O1 | 20.1 (3) | C14—C15—C16—C17 | 1.6 (3) |
| C6—N1—C7—C8 | −154.53 (15) | S2—C15—C16—C17 | −179.32 (17) |
| O1—C7—C8—S1 | 22.1 (2) | C13—C12—C17—C16 | −0.4 (3) |
| N1—C7—C8—S1 | −163.17 (12) | C11—C12—C17—C16 | 178.58 (18) |
| C9—S1—C8—C7 | 90.32 (13) | C15—C16—C17—C12 | −0.8 (3) |
| N3—N2—C9—N4 | −0.06 (17) | C9—N4—C18—C23 | 104.7 (2) |
| N3—N2—C9—S1 | −178.58 (12) | C10—N4—C18—C23 | −85.2 (2) |
| C10—N4—C9—N2 | −0.32 (17) | C9—N4—C18—C19 | −78.7 (2) |
| C18—N4—C9—N2 | 171.54 (14) | C10—N4—C18—C19 | 91.5 (2) |
| C10—N4—C9—S1 | 178.29 (11) | C23—C18—C19—C20 | 0.3 (3) |
| C18—N4—C9—S1 | −9.9 (2) | N4—C18—C19—C20 | −176.41 (18) |
| C8—S1—C9—N2 | −14.31 (16) | C18—C19—C20—C21 | −0.3 (4) |
| C8—S1—C9—N4 | 167.32 (13) | C19—C20—C21—C22 | 0.0 (4) |
| N2—N3—C10—N4 | −0.65 (17) | C20—C21—C22—C23 | 0.4 (4) |
| N2—N3—C10—C11 | 177.73 (14) | C19—C18—C23—C22 | 0.1 (3) |
| C9—N4—C10—N3 | 0.60 (17) | N4—C18—C23—C22 | 176.72 (19) |
| C18—N4—C10—N3 | −171.37 (14) | C21—C22—C23—C18 | −0.4 (4) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1N1···N3i | 0.94 | 2.04 | 2.9787 (19) | 174 |
| C8—H8A···O1ii | 0.97 | 2.52 | 3.147 (2) | 123 |
| C11—H11B···O1iii | 0.97 | 2.47 | 3.416 (2) | 165 |
Symmetry codes: (i) x, −y+1/2, z+1/2; (ii) −x+1, −y+1, −z+2; (iii) −x+1, −y+1, −z+1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: LH5283).
References
- Bruker (2009). APEX2, SAINT and SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Burch, H. A. & Smith, W. O. (1966). J. Med. Chem. 9, 405–408. [DOI] [PubMed]
- Holla, B. S., Poojary, K. N., Rao, B. S. & Shivananda, M. K. (2002). Eur. J. Med. Chem. 37, 511–517. [DOI] [PubMed]
- Holla, B. S., Veerendra, B., Shivananda, M. K. & Poojary, B. (2003). Eur. J. Med. Chem. 38, 759–767. [DOI] [PubMed]
- Kalyoncuoglu, N., Rollas, S., Sür-Altiner, D., Yegenoglu, Y. & Ang, Ö. (1992). Pharmazie, 47, 796–797. [PubMed]
- Mhasalkar, M. Y., Shah, M. H., Nikam, S. T., Anantanarayanan, K. G. & Deliwala, C. V. (1970). J. Med. Chem. 13, 672–674. [DOI] [PubMed]
- Mir, I., Siddiqui, M. T. & Comrie, A. (1970). Tetrahedron, 26, 5235–5238. [DOI] [PubMed]
- Rudnicka, W., Foks, H., Janowiec, M. & Zwolska-Kwiek, Z. (1986). Acta Pol. Pharm. 43, 523–528. [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536811027565/lh5283sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536811027565/lh5283Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536811027565/lh5283Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report