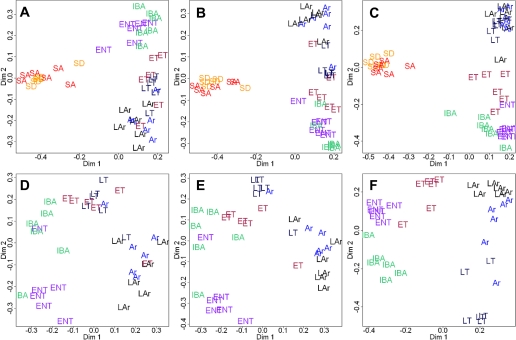
Fig. 5.
Unsupervised classification by Random Forests (50,000 trees) reveals a distinct summer (SA, SD) vs. winter (Ent, ET, LT, Ar, LAr, IBA) switch (A–C) as well as intrawinter cycling (D–F) in the skeletal muscle proteome of hibernating 13-lined ground squirrels (n = 6 per state, see materials and methods). Patterns of clustering were maintained or improved by limiting the data inputs (left to right), ultimately revealing the minimum number of proteins required to define the states. A (all states) and D (winter only) show Random Forests outputs from all completely matched spots (i.e., present on all analytical gels), while B (all) and E (winter) reflect all 133 of those that were identified. C and D: minimum number of known proteins required for equivalent or improved clustering compared with the expanded data inputs. This was accomplished with 10 protein spots for all states, and 12 proteins for winter states only (listed in Table 3).