Abstract
The number of individual cases of psychiatric disorders that can be ascribed to identified, rare, single mutations is increasing with great rapidity. Such mutations can be recapitulated in mice to generate animal models with direct etiological validity. Defining the underlying pathogenic mechanisms will require an experimental and theoretical framework to make the links from mutation to altered behavior in an animal or psychopathology in a human. Here, we discuss key elements of such a framework, including cell type-based phenotyping, developmental trajectories, linking circuit properties at micro and macro scales and definition of neurobiological phenotypes that are directly translatable to humans.
Keywords: autism, schizophrenia, rare mutations, synaptic, interneurons, EEG, functional connectivity, microcircuits, Cre, allelic heterogeneity
Historical approaches to modeling psychiatric disorders
Psychiatric disorders constitute a diverse set of conditions, variously impinging on all domains of mental function and affecting the most fundamental human attributes: language, thought, perception, mood and sense of self. Collectively, they cause a substantial public health burden, greater than cancers or cardiovascular disease [1]. The proportional burden is actually increasing as we learn more about the molecular bases of other common diseases and how to treat them, through the application of molecular genetics. Until recently, these approaches have been difficult in psychiatry as we have had few entry points to the underlying molecular mechanisms. This is now changing as psychiatric genetics reveals more and more specific mutations predisposing to psychiatric disease. These discoveries, along with revolutionary advances in neurogenetic techniques, provide the means to elucidate pathogenic mechanisms through modeling such mutations in animals and defining their effects at the neurobiological level.
In the absence of such information on specific genetic causes, previous modeling approaches for psychiatric disorders have been based either on surface similarities between behavioral assays in rodents and domains of psychopathology in humans or on dissecting the psychopharmacological mechanisms of known drugs [2,3]. These have both centered on the goal of generating relevant behavioral assays that can provide platforms for drug screening.
The behavioral approach first defines behavioral assays in animals that are thought to mimic symptoms in particular psychological domains [4,5]. Thus, 'learned helplessness' is thought to model depressive symptoms, spontaneous alternation or latent inhibition are believed to index working memory, which is often deficient in schizophrenia, and decreased social interactions in rodents are thought to relate to similar effects in humans with autism. Animals can be generated with defects in any of these or similar domains by a variety of manipulations, including pharmacological, surgical, experiential and genetic. The relevance (or predictive validity) of these models to human psychopathology has generally been tested by whether they are responsive to known medications, such as antidepressants or antipsychotics [3]. Using these approaches, it has been possible to generate a large number of such models, which have been used to screen through drug libraries for novel therapeutic compounds or to test prototypes of existing medication. Unfortunately, these approaches have yielded few new drugs [6,7].
There are several drawbacks to using only the criterion of response to known medication. First, most of the models do not have causal etiological validity - acute blockade of NMDA-type receptors for glutamate or ventral hippocampal lesion are not actual causes of schizophrenia, for example. Second, the surface parallels between rodent and human behavior may be misleading. On the contrary, we should expect some of the underlying neurobiological defects to manifest at the behavioral level in species-specific ways, which may or may not appear related [2]. Third, defining predictive validity based on responsiveness to known drugs inevitably restricts the focus to mechanisms affected by those drugs. This circularity has undoubtedly contributed to the paucity of new drug mechanisms being found.
A complementary approach is now becoming possible as particular mutations causing psychiatric illness are being identified at an ever-increasing rate. Mouse models of these mutations can now be investigated using increasingly powerful and sophisticated tools to get at the underlying neurobiology and elucidate pathogenic mechanisms, from the level of synapses formed between specific cell types in specific circuits to the level of dynamic function of neuronal networks and concomitant moment-to-moment behavior. These types of approaches should provide the necessary neurobiological knowledge to enable the rational design of new therapeutics.
A revolution in psychiatric genetics
The field of psychiatric genetics is undergoing a profound paradigm shift. For several decades, the prevailing model has held that psychiatric disorders arise in any individual due to the cumulative effects of a large number of common variants [8-10]. Each of these, by themselves, would have a very small effect on risk, but when the collective burden of such alleles passes a putative threshold, the system would be pushed into a pathogenic state. Though this model has little empirical support, it provided the theoretical foundation for genome-wide association studies (GWAS) [11], which aim to detect such variants by comparing allele frequencies for millions of such variants between cases and controls [12-15]. For psychiatric disorders, these studies, now carried out on tens of thousands of people, have yielded a number of replicated common variants reaching the threshold for genome-wide significance [16-26]. These include eight loci for schizophrenia [26,27], two for bipolar disorder [25] and one for schizophrenia and bipolar disorder together [25]; three potential loci arising from individual GWAS for autism [22-24] have not yet been replicated on the same scale. These findings point to loci that may be involved in disease risk at a population level but do not identify or speak to the likely allelic frequency of causal variants [28]. Each of the associated variants has a tiny statistical effect on risk at the population level and, collectively, the significant single nucleotide polymorphisms statistically account for only a few percent of the overall heritability of the disorder [18,25,26].
The alternative model is that psychiatric disorders arise due to mutations in any of a very large number of genes [11,29-33]. Under this model, psychiatric diagnostic categories are actually umbrella terms for large numbers of distinct genetic disorders that happen to result in similar spectra of symptoms. This is the sort of genetic heterogeneity that underlies categories such as congenital deafness, epilepsy, mental retardation, retinitis pigmentosa, many cancers and other conditions [34,35].
It has, of course, been known for some time that psychiatric illness could arise due to single mutations. Well-known examples include fragile × syndrome [36], Rett syndrome [37] and mutations in the neuroligin genes NLGN3 and NLGN4 [38], all of which are associated with autistic spectrum disorder, and velocardiofacial syndrome (22q11.2 deletion syndrome) [39] and the Scottish DISC1 translocation [40], which are associated with schizophrenia and other psychiatric diagnoses. The number of such identified mutations is now steadily and rapidly increasing, thanks to the application of new genomic microarray [41,42] and sequencing technologies [43,44], to the point where they collectively explain an appreciable fraction of psychiatric diagnoses.
Copy number variants (deletions or duplication of chromosomal segments, often affecting multiple genes) have been most readily identified and make up an important class of causal mutations in schizophrenia [42,45-51], autism [41,52-54], attention deficit-hyperactivity disorder [55-58], Tourette syndrome [59], developmental delay and mental retardation [60,61], epilepsy [62] and cortical malformations [63]. Whole-exome and whole-genome sequencing approaches are now also identifying large numbers of point mutations individually responsible for psychiatric conditions [64-71].
A number of important principles have emerged from these studies. First, there is considerable overlap in the genetic etiology of what had previously been considered distinct disorders. Individual mutations that predispose to one class of psychiatric illness, such as schizophrenia, are also associated with other disorders, such as bipolar disorder, autism, mental retardation, epilepsy, attention deficit hyperactivity disorder and Tourette's syndrome (for example, [31,72-77]), in agreement with recent epidemiological data indicating shared risk [78-83]. Traditional diagnostic categories, although still very useful in organizing daily practice in psychiatry, may therefore represent not natural kinds in terms of etiology, but more or less distinct phenotypic endpoints that may arise from common origins.
Second, the mutations so far discovered are characterized by incomplete penetrance and variable expressivity [31,77,84]. As with the DISC1 translocation, such mutations may result in a range of phenotypes and many carriers may be unaffected by any psychiatric condition [85]. Of course, the penetrance depends on which phenotype is being assessed - it will be lowest for specific diagnoses, higher for psychiatric illness generally and higher still for neurobiological endophenotypes, which may be apparent even in clinically unaffected carriers.
Third, many of the identified genes are involved in neural development [42,77,86]. While certainly not exclusive, this is probably the largest category of susceptibility genes. Genes involved in activity-dependent synaptic plasticity, such as FMR1, are also highly represented. With increasing numbers of genes being identified all the time, it is becoming possible to assign many of them to specific biochemical pathways or cellular processes, such as synapse formation and plasticity (for example, [64,66,87-89]).
Fourth, allelic specificity and dosage are extremely important. Different mutations in the same gene may result in very different phenotypic outcomes. As a classic example, Duchenne muscular dystrophy and Becker's muscular dystrophy are caused by different types of mutations in the dystrophin gene: their clinical severity, manifestation, and age of onset are also different [90]. Similar effects are seen for genes implicated in neurological and psychiatric disorders [32,35], as described later. In addition, some alleles may cause severe neurological disorders when homozygous but manifest as psychiatric illness in heterozygous form [32,35,91].
Fifth, while these findings highlight the importance of rare single mutations, they do not necessarily imply a simple mode of inheritance. Many of the mutations found so far show a dominant effect, but recessive mutations are likely to also contribute a sizeable fraction of cases [71,92]. In addition, there is likely to be an important role for modifying mutations in the genetic background that can alter the phenotypic expression of the 'primary' mutation. This is the norm, even for the most classically 'Mendelian' disorders [35,77,93]. One should thus expect a distribution of genetic mechanisms across cases - some will be caused by highly penetrant mutations, others by mutations with more variable outcome, which are modified to some extent by the genetic background, and yet others will involve the inheritance of two or more distinct mutations [35,77,94-96].
Sixth, the eventual phenotype will also be modified by non-genetic factors, including (i) intrinsic developmental variation, where the phenotypic outcome varies due to inherent noise in the molecular processes mediating neurodevelopment [97], and (ii) environmental risk factors, which have been implicated by epidemiological studies. For schizophrenia, for example, these include maternal infection, urbanicity, migration and cannabis use [98]. The impact of such factors in individuals may be highly uneven and dependent on genotype.
Despite these complexities, the major finding is clear: mutations with large effect on risk of psychiatric disorders exist and we now have the means to identify them. If we think of this as a genetic screen for mutations causing a specific phenotype, with demonstrated saturation mutagenesis of the human population [32], then the most effective approach to follow up these discoveries is clear: find the mutations of largest effect and use these as entry points to elucidate the underlying biology.
Bone fide models of genetic etiology
Advances in genetic engineering allow us to recapitulate human mutations in mice, and increasingly in rat. It is possible to generate animals with full gene knock-outs, conditional removal of the gene with spatial and temporal control, knock-in of exact human alleles, and to precisely engineer genomic deletions or duplications into syntenic regions of mouse chromosomes, thereby yielding animal models with direct construct validity; that is, where the manipulation is an actual cause of the condition in humans [99,100]. An emerging principle is the importance of allelic specificity for phenotypic outcome. For example, mouse mutants of Nlgn3 and Shank3 that recapitulate human alleles found in autism patients demonstrate selective effects that differ from null alleles [101-103]. Mice modeling human deletions or duplications can also be generated [104-106], and phenotypically compared with mutations of single genes in the affected regions in order to track down the specific culprits [107,108].
The number of models generated to date that directly recapitulate or mimic the effects of human alleles is small, but they have already begun to reveal details of the mechanisms of pathogenesis in specific mutants and to highlight convergent pathways, as well as some important general principles. For example, mutations in Nlgn, Nrxn (encoding a neurexin), Shank3 (encoding an adaptor protein that interacts with the cytoplasmic tails of Nlgn proteins) and Cntnap2 (which also encodes a member of the neurexin protein family) all affect synapse formation, altering the biochemical composition of synapses and the balance between excitation and inhibition between specific cell types in developing neuronal networks, with a range of concomitant behavioral deficits [101-103,109-112]. Local and long-range circuitry can also be altered by defects in cell migration and axon projection, which have been observed in DISC1 mutants or transgenic lines (for example, [113,114]), paralleling observations in humans with DISC1 mutations [115]. Alterations in long-range functional connectivity have also been observed - for example, between cortex and striatum in Shank3 mutant mice [111], between cortex and hippocampus in Df(16)A+/- mice, which recapitulate 22q11.2 deletion [105], and in cortical synchrony in Cntnap2 mutant mice [112]. These kinds of observations help generate specific hypotheses regarding the cellular origins of pathophysiological states, which can now be tested with the increasingly sophisticated neurogenetic tools available (for example, [116-118]).
Studies in some animal models, such as mice mutant in Fmr1 or MeCP2 (the gene for methyl CpG binding protein 2, mutated in Rett syndrome), have even suggested possible new therapies based on detailed understanding of the biochemical pathology [119-121]. Despite all these advances, however, the complete pathways of pathogenesis for any single case remain obscure. Even for Fmr1, where the effects of mutation of this gene on synaptic plasticity are well described at the biochemical and cellular levels, the ultimate impact of these changes on the functions of specific neural circuits and how these explain the observed behaviors or psychopathology are only beginning to be unraveled [122].
To fully elucidate the pathogenic mechanisms for any specific mutation will require the integration of analyses across multiple levels, from protein function to systems neuroscience. What are the biochemical functions of the mutated protein? What are the cellular effects in various brain regions? How do these affect the function of various neural circuits? How do changes in neural activity play out over subsequent activity-dependent development? What are the cascading or reactive consequences as the brain develops with these initial alterations? What are the concomitant effects on physiology and behavior? Which circuit defects are responsible for which behavioral phenotypes? Which are the ones most relevant to the symptoms of the human disease? Which are the ones most amenable to intervention? These are formidable challenges even in light of the spectacular advances in modern neuroscience.
Rather than predefining which areas or mechanisms are important or how we expect animal models of psychiatric disorders to behave, we can take a bottom-up approach and simply ask, for any of these mouse mutants 'what happens to their brain circuits?' In analyzing the effects of any particular mutation it will be essential to consider the full cellular complexity of the circuitry and to use all the tools of modern molecular neuroscience to dissect the causal chains from primary genetic lesion to ultimate phenotype(s).
A cell type-based approach is key to link genes and circuits in discovering pathogenic mechanisms
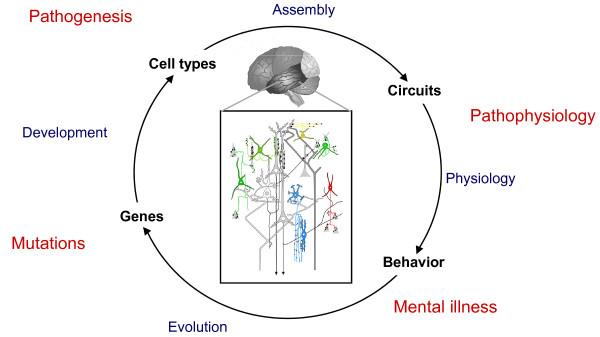
Is there a generally useful strategy to tackle the problem of circuit pathogenesis in rodent models? If genes are the entry points to the cause and genetic architecture of psychiatric disorder, then we suggest that specific cell types may be the next set of entry points in neurobiology studies that may coherently link different levels of analysis, integrate different approaches and technologies, and yield useful answers in understanding pathogenic mechanisms. This is based on the following three first principles: individual cell types are the basic components of neural circuit organization, the building blocks of circuit assembly, and the basic units of gene expression and regulation in the brain (Figure 1). Therefore, a research program centered around cell type-based approaches is likely to be most productive in discovering the biological processes that build and operate neural circuits, and in revealing how these processes are altered by mutations and environmental insults.
Figure 1.
A framework to elucidate pathogenic mechanisms from mutations to mental illness. The effects of mutations in different genes can be analyzed across development on diverse cell types, local circuit organization and emergent properties, functional connectivity on a macro scale and correlated behavior in animals or psychopathology in humans.
The focus on cell types embraces the importance of different types of synapse. For example, mutations in Nlgn genes differentially affect the activity-dependent stabilization of excitatory (Nlgn1) or inhibitory (Nlgn2) synapses, selectively altering levels of different neurotransmitter receptors [123-125]. The effect of Nlgn2 mutation is specific for inhibitory synapses from fast-spiking (parvalbumin-positive) but not from somatostatin-positive interneurons [126]. Such effects can also be region-specific: Nlgn3 knock-in mice show increased synaptic inhibition in the cortex, but increased excitatory synaptic transmission in the hippocampus [102]. Mutations in other genes, such as Fmr1 [122] and ErbB4 or Nrg1 [127], show similarly cell type- and synapse type-specific effects. Understanding the particular profile of effects across multiple cell types in each mutant thus requires engagement with the cellular complexity of these circuits. This profile will also importantly include compensatory changes that may be secondarily induced and that could play important roles in pathogenesis (for example, [128-131]).
However, cell type-based analysis of neural circuits, although more routinely practiced in invertebrate systems, is highly challenging in the mammalian brain circuits, which consist of highly diverse and intermingled cell types that, until recently, could not be readily identified and experimentally accessed.
Fortunately, our ability to carry out such cell type-based analyses is rapidly increasing, as new neurogenetic tools are developed. Gene expression profiles that define many cell types have been elucidated, enabling researchers to isolate specific genetic elements that can be used to drive cell type-specific expression of a variety of transgenic constructs with different uses. These include tracing axonal projections and synaptic connections, monitoring electrical activity, and non-invasively activating or shutting off neurons in behaving animals with optical methods or chemical ligands [117,132,133]. Combined with advances in multi-photon imaging, automated microscopy and computational resources to deal with very large datasets, these tools confer unprecedented power to dissect the structure and function of neuronal circuits [134,135].
The Cre-loxP system has been adopted as a standardized approach in the mouse to drive such constructs in specific cell types [136]. This binary gene expression system is an effective strategy that confers cell type specificity as well as combinatorial power. A Cre driver mouse line provides a 'genetic switch' in a cell type that, through site-specific recombination between loxP recognition sites, can turn on or off the various molecular probes that allow these cells to be analyzed by all the above modern techniques in their native circuit in vivo. An increasing number of Cre drivers thus will allow more systematic and integrated analysis of different cell types in the relevant circuits (for example, [137,138]). Furthermore, certain Cre drivers allow tracking the developmental history of cell types, thereby enabling a comprehensive analysis of these cell types, from their specification, migration and synapse development to their integration and function in the circuits. This approach will be particularly powerful in revealing how the developmental trajectories of cells and circuits are altered, what are the primary cell autonomous changes due to 'direct hit' of the mutant gene, and what are the secondary, possibly maladaptive changes that lead to aberrant circuit-level operations. Genetic tracking of the development of distinct cell types thus will begin to link studies of circuit assembly and function and their alterations in disease models. This can be achieved simply by bringing the Cre driver allele and relevant reporter alleles into an etiology model through breeding.
Analysis of multiple circuits brain-wide in a disease model will allow a more unbiased 'screen' for deficits, less restricted by prior knowledge and assumptions of disease pathology. This will allow definition of the overall profile of deficits across multiple systems in each individual mutant, which may be quite unique. Comparison of multiple disease models promises to identify shared or distinct circuit deficits in these models, which may underlie their shared or distinct behavioral phenotypes. To achieve this, it will be increasingly useful to establish high-throughput methods to effectively screen for alterations in cell numbers, connectivity, and function with cell type resolution [134].
The convergence of phenotype across multiple animal models may arise at the biochemical or cellular level in some cases (such as pathways involved in synapse formation [87,88] or in cAMP signaling [139]), but only at the level of the emergent functions of large-scale neuronal networks in others (such as the responsiveness of the dopaminergic system [129,140] or GABAergic signaling and neuronal synchrony in the prefrontal cortex [131]). A central challenge in this framework is thus to link the cellular level analysis of phenotypic effects on microcircuit architecture to the functional consequences for brain function on a larger scale. It is these emergent functions that will best explain behavior or psychopathology. Rather than merely demonstrating that a mouse mutant shows some patterns of behavior reminiscent of human symptomatology, it will be essential to analyze physiological measures of circuit dynamics that explain behavior on a moment-to-moment basis in the relevant social and cognitive context.
Large-scale analysis of circuits that subserve behavior
The neuroscience of animal and human behavior has witnessed a productive paradigm shift in the past decade where the trend of assigning complex traits to single genes has given way to the realization that dynamic coordination between micro and macro circuits is key to understanding the neuronal basis of behavior [141]. In other words, the emphasis has shifted from associating a behavior to a single gene or its protein product simply because mutation of that gene disrupts that behavior to understanding how functional variability at the gene or protein level influences the dynamic coordination of neurons in systems that support a certain behavior.
In the context of schizophrenia, the approach has been descriptive and mechanistically naive in that, for the most part, it has focused on associating a common allelic variation, which in some cases has not even been associated with a functional alteration, to complex behavioral disruptions whose physiology is still not fully understood. It is now increasingly being appreciated that a mechanistic understanding of the pathophysiology of schizophrenia is contingent upon (i) a better understanding of the dynamic coordination of neuronal processes that serve the behaviors that are disturbed in at least some cases of schizophrenia and (ii) understanding the functional role of implicated genes in micro and macro neuronal circuits that subserve this physiology. Recent studies, highlighted below, are beginning to make considerable advances in these two approaches.
Electrophysiological recordings using electroencephalography (EEG) have for decades reported behaviorally relevant oscillatory activity throughout the cerebral cortex [142]. Until recently, however, these data did not receive much attention in the context of psychiatric disorders, mostly because these measures had poor spatial and temporal resolution and were therefore considered functionally vague. The advent of magnetoencephalography (MEG) and development of better EEG instrumentation combined with invasive electrophysiological recordings in laboratory animals has now provided strong evidence that these measures are biologically meaningful correlates of behavior [143]. This has been in part because of a series of elegant studies in animals establishing a relationship between single neuron activity and local field potential oscillations in behaviorally relevant contexts [144,145].
Animal studies have further demonstrated that synchronized neuronal oscillations at various frequencies are a measure of coordinated neuronal activity that supports behavior. EEG and MEG studies in humans have subsequently established a close relationship between oscillations at various frequencies and behavioral performance, including working memory and selective attention [146]. This has been followed by several studies reporting changes in neuronal oscillations associated with cognitive deficits as well as some symptoms of schizophrenia [147-150]. EEG and MEG are thus promising translational methods that assess oscillatory activity with similar temporal and spatial resolution to more invasive methods that can be used in behaving animals, as described later.
Thus, experimental procedures such as pharmacological and genetic manipulations can be applied in conjunction with single unit recording and local field potentials to understand the molecular and cellular processes that could contribute to abnormal oscillatory activities [151,152]. The emergent properties of neuronal networks depend crucially on the cellular architecture of the microcircuits involved and on the interactions between definable cell types. For example, the connectivity between parvalbumin-positive interneurons and pyramidal neurons is essential to drive oscillations in the gamma frequency, for phase-locking between rhythms at different frequencies and for long-range temporal coherence between brain areas, which subserves cognitive and perceptual functions [116,151,153-157]. Other interneuron subtypes have dissociable functions on neural systems [158-161].
The proliferation of cell type-specific genetic drivers and development of optogenetic approaches also enable follow-up experiments to directly test hypotheses arising from studies of mutant phenotypes by manipulating the activity of the implicated cell types in highly specific ways [117]. Such approaches should allow the field to move beyond correlations between various anatomical and physiological disturbances in mutant animals to direct tests of how the precise connectivity patterns and strengths between different cell types affect the information-processing parameters of microcircuits and larger neuronal networks and impact on specific behaviors [116,118,154,162].
While mice are likely to continue to lead the way in the discovery of the effects of specific mutations on nervous system development and function, we can also expect the increasing use of genetically manipulated rat and primate models to further dissect pathophysiological mechanisms. Zinc-finger nucleases can be used to specifically modify the genome in effectively any type of cell, including embryonic stem cells or early embryos, thus generating genetically modified organisms [163]. This approach has been used to generate gene knockouts [164] and also to knock in specific alleles in the rat [165,166], opening up the possibility of recapitulating human alleles in this species and others in the future. Brain cells can also be genetically modified in a cell type-specific manner by viral transfection [132,167], making them accessible to acute gene manipulations and also to optogenetic techniques in any species, including rats and non-human primates [168,169]. These species, which have a much richer tradition of systems neuroscience and greater cognitive sophistication than the mouse, will no doubt be essential in further elucidating general principles and mechanisms of pathophysiology and relating them to humans.
Translation to humans
Because diagnostic classification of mental illnesses in clinical psychiatry at present is merely based on phenomenology and not disease etiology, it is important to consider translation between humans and rodent models not via clinical diagnosis but specific biological traits. These traits would include behavioral constructs and physiological characteristics, and are crucial in understanding disease mechanisms in rodent models. Even more importantly, such characteristics can be used as indicators of drug screening in rodents and markers in clinical trials with humans.
A major confound in analyzing neurobiological intermediate phenotypes in psychiatric patients has been the extreme underlying (and previously cryptic) genetic heterogeneity. As the genetic factors involved in psychiatric illness are determined in more and more patients, it should be possible to make much more direct and powerful comparisons of underlying phenotypic traits between genetically defined subsets of humans and mice carrying the same mutation. This genetic stratification of patients may also prove invaluable in the design of clinical trials aimed at primary causes. On the other hand, the analysis of phenotypes correlated with clinical states will need to look for convergence across mouse mutants or human patients with diverse genetic causes.
Many efforts have been made to try to identify equivalent characteristics between humans and rodents. For example, at the behavioral level, the Cognitive Neuroscience Treatment Research to Improve Cognition in Schizophrenia (CNTRICS) aims to develop and implement animal model paradigms that can tap the cognitive and emotional-processing constructs translatable to humans [170]. Although only limited numbers of paradigms are widely accepted to be translational, collaboration among human neuropsychologists and animal neurobehavioral scientists may identify several paradigms to be utilized for translation. Starting these studies with animal models with direct etiological validity should tell us which behavioral constructs are really most relevant to human psychopathology, even if the effects manifest differently on the surface.
Given well-founded concerns over the species-specificity of behavioral measures, it seems likely that neurophysiological measures may prove to be more translatable between animals and humans. This may be especially true of measures of functional connectivity or coherence between brain areas, as these may most powerfully index differences in network function that affect psychopathology. These can be assayed with EEG and MEG, as described above, but also with functional connectivity studies using functional magnetic resonance imaging to examine correlations between activities of remote but connected regions [171-174]. Although these measures are simply about observed correlations, they provide clinically relevant dynamic information that can be invaluable for animal studies, which can in turn provide more detailed mechanistic information about the neuronal basis of the reported aberrant connectivity. For example, the reduced synchrony between hippocampal and prefrontal regions in the 22q11.2 deletion Df(16)A+/- mice [105] parallels observations in humans with schizophrenia [171], generating testable hypotheses about the origins of this defect [157]. Alterations in cortico-striatal or intracortical coherence in Shank3 [111], Fmr1 [122] and Cntnap2 mutant mice [112] similarly provide entry points to define the pathophysiology of these disorders at a level that can be directly assessed in humans, especially those with mutations in the same genes.
Definition of the mouse correlates of human pathophysiology and psychopathology will also allow a reverse translational approach to identify new candidate susceptibility genes. Prospective phenotyping of mice with mutations in neurodevelopmental genes may reveal phenotypes paralleling established models (for example, NogoA [175], Dlg4 [176], Sema6A [177] and Slitrk5 [178]). This gives a higher prior probability for assigning pathogenicity in the event that mutations in those genes are then found in human patients.
Strategies towards new treatments
The drugs currently used to treat disorders like schizophrenia were discovered serendipitously, are only partially effective for positive symptoms but not the more debilitating negative and cognitive symptoms, work in some patients and not others and have serious side effects [179]. For autism spectrum disorder, the situation is even more bleak - while there are some drugs prescribed to treat some symptoms there is no overall therapeutic of proven value [180]. The main reason hardly any new drugs (with novel mechanisms) have been successfully developed in the past 60 years has been our lack of understanding of the underlying mechanisms of these disorders.
New genetic models may be utilized in identifying novel treatment strategies in three ways: first, to identify novel targets through which we can find more effective and safer compounds for treatment within the current understanding of these diseases; second, to identify novel targets and compounds that may not be obvious from current paradigms; third, and probably most important, to identify targets and compounds for early intervention.
By analyzing molecular, cellular and circuitry disturbances in new genetic models, we may find better clues to how the cellular pathways involving the gene of interest may interact with currently accepted pathophysiological paradigms (such as the roles of GABA, NMDA receptors and dopamine receptors in schizophrenia). In such cases, we may identify better drug targets, against which we can expect compounds with better efficacy and less side effects.
However, our hope is to identify new targets and compounds with new genetic models. Although investigators who generate new genetic models tend to test whether current medications (such as clozapine and haloperidol for schizophrenia) can normalize deficits in behavioral paradigms, it may be more advantageous for new drug discovery to identify paradigms that are relevant to human diseases (that is, likely to correlate not to one disease but to several), but that cannot be normalized by current medications. For example, a DISC1 transgenic mouse model with neonatal poly(I:C) treatment (to mimic maternal infection) shows many types of behavioral deficit, most of which are normalized by administration of clozapine [181]. However, the impairment of social behaviors in this model is resistant to both clozapine and haloperidol treatment. Provided that these impairments are, at least in part, relevant to negative symptoms, screening of novel compounds to normalize this behavior in this model may lead to the identification of new treatments for negative symptoms.
Early intervention is now becoming a key in treatment strategies in many brain disorders, such as Alzheimer's disease and schizophrenia. Animal models, especially those in which neurodevelopmentally important genes are targeted, are very useful to dissect the pathological course even from the premorbid stage to the full onset. For some symptoms, specific common phenotypes might be due to secondary mechanisms by which the developing brain reacts to diverse primary insults. For example, altered dopaminergic signaling may arise as a result of a variety of primary defects and may represent a final common pathway to psychosis [129,130,140]. The molecular homeostatic mechanisms underlying these secondary changes may thus represent a viable target for early intervention in genetically at-risk subjects. Similar events may underlie the emergence of dysfunction in other circuits across other disorders. In the case of schizophrenia where the intervention has to occur during early to late adolescence, the major obstacle to moving intervention to an earlier age is concerns over the effects of manipulating brain systems that are still developing. These concerns are compounded by a lack of concrete understanding of the biology of adolescent brain, especially in the context of the physiology of affect and cognition. Animal models will, therefore, be critical in advancing this aspect of the field.
A way forward
The discovery of mutations that strongly predispose individuals to psychopathology provides a crucial starting point to define pathogenic mechanisms and pathways. Direct animal models of genetic etiology can be analyzed in a comprehensive and systematic way, using the full arsenal of modern neuroscience to link effects on particular cell types in defined microcircuits to emergent properties of larger scale brain networks in behaviorally relevant contexts. These analyses will hopefully reveal neurobiological phenotypes that can be translated very directly to humans, suggest points of possible therapeutic intervention and aid the rational design of new drugs targeted at the underlying causes of mental illness.
Contributor Information
Kevin J Mitchell, Email: Kevin.Mitchell@tcd.ie.
Z Josh Huang, Email: huangj@cshl.edu.
Bita Moghaddam, Email: bita@pitt.edu.
Akira Sawa, Email: asawa1@jhmi.edu.
References
- World Health Organisation. The Global Burden of Disease. 2004 Update. Geneva: WHO; 2008. [Google Scholar]
- Insel TR. From animal models to model animals. Biol Psychiatry. 2007;62:1337–1339. doi: 10.1016/j.biopsych.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Moore H. The role of rodent models in the discovery of new treatments for schizophrenia: updating our strategy. Schizophr Bull. 2010;36:1066–1072. doi: 10.1093/schbul/sbq106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell CM, Miyakawa T. Schizophrenia-relevant behavioral testing in rodent models: a uniquely human disorder? Biol Psychiatry. 2006;59:1198–1207. doi: 10.1016/j.biopsych.2006.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker M. Animal models: inside the minds of mice and men. Nature. 2011;475:123–128. doi: 10.1038/475123a. [DOI] [PubMed] [Google Scholar]
- Moghaddam B, Javitt D. From revolution to evolution: the glutamate hypothesis of schizophrenia and its implication for treatment. Neuropsychopharmacology. 2011. in press . [DOI] [PMC free article] [PubMed]
- De Fruyt J, Deschepper E, Audenaert K, Constant E, Floris M, Pitchot W, Sienaert P, Souery D, Claes S. Second generation antipsychotics in the treatment of bipolar depression: a systematic review and meta-analysis. J Psychopharmacol. 2011. in press . [DOI] [PubMed]
- Gottesman II, Shields J. A polygenic theory of schizophrenia. Proc Natl Acad Sci USA. 1967;58:199–205. doi: 10.1073/pnas.58.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N. Genetic linkage and complex diseases, with special reference to psychiatric disorders. Genet Epidemiol. 1990;7:3–16. doi: 10.1002/gepi.1370070103. discussion 17-45. [DOI] [PubMed] [Google Scholar]
- Wray NR, Visscher PM. Narrowing the boundaries of the genetic architecture of schizophrenia. Schizophr Bull. 2010;36:14–23. doi: 10.1093/schbul/sbp137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell KJ, Porteous DJ. Rethinking the genetic architecture of schizophrenia. Psychol Med. 2011;41:19–32. doi: 10.1017/S003329171000070X. [DOI] [PubMed] [Google Scholar]
- Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- Reich DE, Lander ES. On the allelic spectrum of human disease. Trends Genet. 2001;17:502–510. doi: 10.1016/s0168-9525(01)02410-6. [DOI] [PubMed] [Google Scholar]
- Psychiatric Genetics Consortium. A framework for interpreting genome-wide association studies of psychiatric disorders. Mol Psychiatry. 2009;14:10–17. doi: 10.1038/mp.2008.126. [DOI] [PubMed] [Google Scholar]
- Sullivan PF. The psychiatric GWAS consortium: big science comes to psychiatry. Neuron. 2010;68:182–186. doi: 10.1016/j.neuron.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah S, Nelson MR, Delisi LE, Reneland RH, Markward N, James MR, Nyholt DR, Hayward N, Handoko H, Mowry B, Kammerer S, Braun A. Identification of the semaphorin receptor PLXNA2 as a candidate for susceptibility to schizophrenia. Mol Psychiatry. 2006;11:471–478. doi: 10.1038/sj.mp.4001785. [DOI] [PubMed] [Google Scholar]
- O'Donovan MC, Craddock N, Norton N, Williams H, Peirce T, Moskvina V, Nikolov I, Hamshere M, Carroll L, Georgieva L, Dwyer S, Holmans P, Marchini JL, Spencer CC, Howie B, Leung HT, Hartmann AM, Möller HJ, Morris DW, Shi Y, Feng G, Hoffmann P, Propping P, Vasilescu C, Maier W, Rietschel M, Zammit S, Schumacher J, Quinn EM, Schulze TG. et al. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat Genet. 2008;40:1053–1055. doi: 10.1038/ng.201. [DOI] [PubMed] [Google Scholar]
- Purcell SM, Wray NR, Stone JL, Visscher PM, O'Donovan MC, Sullivan PF, Sklar P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Need AC, Ge D, Weale ME, Maia J, Feng S, Heinzen EL, Shianna KV, Yoon W, Kasperaviciūte D, Gennarelli M, Strittmatter WJ, Bonvicini C, Rossi G, Jayathilake K, Cola PA, McEvoy JP, Keefe RS, Fisher EM, St Jean PL, Giegling I, Hartmann AM, Möller HJ, Ruppert A, Fraser G, Crombie C, Middleton LT, St Clair D, Roses AD, Muglia P, Francks C. et al. A genome-wide investigation of SNPs and CNVs in schizophrenia. PLoS Genet. 2009;5:e1000373. doi: 10.1371/journal.pgen.1000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H, Ophoff RA, Steinberg S, Andreassen OA, Cichon S, Rujescu D, Werge T, Pietiläinen OP, Mors O, Mortensen PB, Sigurdsson E, Gustafsson O, Nyegaard M, Tuulio-Henriksson A, Ingason A, Hansen T, Suvisaari J, Lonnqvist J, Paunio T, Børglum AD, Hartmann A, Fink-Jensen A, Nordentoft M, Hougaard D, Norgaard-Pedersen B, Böttcher Y, Olesen J, Breuer R, Möller HJ, Giegling I. et al. Common variants conferring risk of schizophrenia. Nature. 2009;460:744–747. doi: 10.1038/nature08186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Levinson DF, Duan J, Sanders AR, Zheng Y, Pe'er I, Dudbridge F, Holmans PA, Whittemore AS, Mowry BJ, Olincy A, Amin F, Cloninger CR, Silverman JM, Buccola NG, Byerley WF, Black DW, Crowe RR, Oksenberg JR, Mirel DB, Kendler KS, Freedman R, Gejman PV. Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature. 2009;460:753–757. doi: 10.1038/nature08192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss LA, Arking DE, Daly MJ, Chakravarti A. A genome-wide linkage and association scan reveals novel loci for autism. Nature. 2009;461:802–808. doi: 10.1038/nature08490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Zhang H, Ma D, Bucan M, Glessner JT, Abrahams BS, Salyakina D, Imielinski M, Bradfield JP, Sleiman PM, Kim CE, Hou C, Frackelton E, Chiavacci R, Takahashi N, Sakurai T, Rappaport E, Lajonchere CM, Munson J, Estes A, Korvatska O, Piven J, Sonnenblick LI, Alvarez Retuerto AI, Herman EI, Dong H, Hutman T, Sigman M, Ozonoff S, Klin A. et al. Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature. 2009;459:528–533. doi: 10.1038/nature07999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anney R, Klei L, Pinto D, Regan R, Conroy J, Magalhaes TR, Correia C, Abrahams BS, Sykes N, Pagnamenta AT, Almeida J, Bacchelli E, Bailey AJ, Baird G, Battaglia A, Berney T, Bolshakova N, Bölte S, Bolton PF, Bourgeron T, Brennan S, Brian J, Carson AR, Casallo G, Casey J, Chu SH, Cochrane L, Corsello C, Crawford EL, Crossett A. et al. A genome-wide scan for common alleles affecting risk for autism. Hum Mol Genet. 2010;19:4072–4082. doi: 10.1093/hmg/ddq307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psychiatric GWAS Consortium Bipolar Disorder Working Group. Sklar P, Ripke S, Scott LJ, Andreassen OA, Cichon S, Craddock N, Edenberg HJ, Nurnberger JI Jr, Rietschel M, Blackwood D, Corvin A, Flickinger M, Guan W, Mattingsdal M, McQuillin A, Kwan P, Wienker TF, Daly M, Dudbridge F, Holmans PA, Lin D, Burmeister M, Greenwood TA, Hamshere ML, Muglia P, Smith EN, Zandi PP, Nievergelt CM, McKinney R, Shilling PD. et al. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet. 2011;43:977–983. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium. Ripke S, Sanders AR, Kendler KS, Levinson DF, Sklar P, Holmans PA, Lin DY, Duan J, Ophoff RA, Andreassen OA, Scolnick E, Cichon S, St Clair D, Corvin A, Gurling H, Werge T, Rujescu D, Blackwood DH, Pato CN, Malhotra AK, Purcell S, Dudbridge F, Neale BM, Rossin L, Visscher PM, Posthuma D, Ruderfer DM, Fanous A, Stefansson H, Steinberg S. et al. Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams HJ, Norton N, Dwyer S, Moskvina V, Nikolov I, Carroll L, Georgieva L, Williams NM, Morris DW, Quinn EM, Giegling I, Ikeda M, Wood J, Lencz T, Hultman C, Lichtenstein P, Thiselton D, Maher BS. Molecular Genetics of Schizophrenia Collaboration (MGS) International Schizophrenia Consortium (ISC), SGENE-plus, GROUP. Malhotra AK, Riley B, Kendler KS, Gill M, Sullivan P, Sklar P, Purcell S, Nimgaonkar VL, Kirov G, Holmans P, Corvin A, Rujescu D. et al. Fine mapping of ZNF804A and genome-wide significant evidence for its involvement in schizophrenia and bipolar disorder. Mol Psychiatry. 2011;16:429–441. doi: 10.1038/mp.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson SP, Wang K, Krantz I, Hakonarson H, Goldstein DB. Rare variants create synthetic genome-wide associations. PLoS Biol. 2010;8:e1000294. doi: 10.1371/journal.pbio.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller MC, Miller G. Resolving the paradox of common, harmful, heritable mental disorders: which evolutionary genetic models work best? Behav Brain Sci. 2006;29:385–404. doi: 10.1017/S0140525X06009095. discussion 405-352. [DOI] [PubMed] [Google Scholar]
- McClellan JM, Susser E, King MC. Schizophrenia: a common disease caused by multiple rare alleles. Br J Psychiatry. 2007;190:194–199. doi: 10.1192/bjp.bp.106.025585. [DOI] [PubMed] [Google Scholar]
- Sebat J, Levy DL, McCarthy SE. Rare structural variants in schizophrenia: one disorder, multiple mutations; one mutation, multiple disorders. Trends Genet. 2009;25:528–535. doi: 10.1016/j.tig.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh CA, Engle EC. Allelic diversity in human developmental neurogenetics: insights into biology and disease. Neuron. 2010;68:245–253. doi: 10.1016/j.neuron.2010.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancur C. Etiological heterogeneity in autism spectrum disorders: More than 100 genetic and genomic disorders and still counting. Brain Res. 2011;1380:42–77. doi: 10.1016/j.brainres.2010.11.078. [DOI] [PubMed] [Google Scholar]
- McClellan J, King MC. Genetic heterogeneity in human disease. Cell. 2010;141:210–217. doi: 10.1016/j.cell.2010.03.032. [DOI] [PubMed] [Google Scholar]
- Lupski JR, Belmont JW, Boerwinkle E, Gibbs RA. Clan genomics and the complex architecture of human disease. Cell. 2011;147:32–43. doi: 10.1016/j.cell.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown WT, Friedman E, Jenkins EC, Brooks J, Wisniewski K, Raguthu S, French JH. Association of fragile × syndrome with autism. Lancet. 1982;1:100. doi: 10.1016/s0140-6736(82)90231-8. [DOI] [PubMed] [Google Scholar]
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- Jamain S, Quach H, Betancur C, Råstam M, Colineaux C, Gillberg IC, Soderstrom H, Giros B, Leboyer M, Gillberg C, Bourgeron T. Paris Autism Research International Sibpair Study. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat Genet. 2003;34:27–29. doi: 10.1038/ng1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karayiorgou M, Morris MA, Morrow B, Shprintzen RJ, Goldberg R, Borrow J, Gos A, Nestadt G, Wolyniec PS, Lasseter VK, Eisen H, Childs B, Kazazian HH, Kucherlapati R, Antonarakis SE, Pulver AE, Housman DE. Schizophrenia susceptibility associated with interstitial deletions of chromosome 22q11. Proc Natl Acad Sci USA. 1995;92:7612–7616. doi: 10.1073/pnas.92.17.7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar JK, Wilson-Annan JC, Anderson S, Christie S, Taylor MS, Semple CA, Devon RS, St Clair DM, Muir WJ, Blackwood DH, Porteous DJ. Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum Mol Genet. 2000;9:1415–1423. doi: 10.1093/hmg/9.9.1415. [DOI] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, Leotta A, Pai D, Zhang R, Lee YH, Hicks J, Spence SJ, Lee AT, Puura K, Lehtimäki T, Ledbetter D, Gregersen PK, Bregman J, Sutcliffe JS, Jobanputra V, Chung W, Warburton D, King MC, Skuse D, Geschwind DH, Gilliam TC. et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, Nord AS, Kusenda M, Malhotra D, Bhandari A, Stray SM, Rippey CF, Roccanova P, Makarov V, Lakshmi B, Findling RL, Sikich L, Stromberg T, Merriman B, Gogtay N, Butler P, Eckstrand K, Noory L, Gochman P, Long R, Chen Z, Davis S, Baker C, Eichler EE, Meltzer PS. et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- Cirulli ET, Goldstein DB. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet. 2010;11:415–425. doi: 10.1038/nrg2779. [DOI] [PubMed] [Google Scholar]
- Cooper GM, Shendure J. Needles in stacks of needles: finding disease-causal variants in a wealth of genomic data. Nat Rev Genet. 2011;12:628–640. doi: 10.1038/nrg3046. [DOI] [PubMed] [Google Scholar]
- Xu B, Roos JL, Levy S, van Rensburg EJ, Gogos JA, Karayiorgou M. Strong association of de novo copy number mutations with sporadic schizophrenia. Nat Genet. 2008;40:880–885. doi: 10.1038/ng.162. [DOI] [PubMed] [Google Scholar]
- Stefansson H, Rujescu D, Cichon S, Pietiläinen OP, Ingason A, Steinberg S, Fossdal R, Sigurdsson E, Sigmundsson T, Buizer-Voskamp JE, Hansen T, Jakobsen KD, Muglia P, Francks C, Matthews PM, Gylfason A, Halldorsson BV, Gudbjartsson D, Thorgeirsson TE, Sigurdsson A, Jonasdottir A, Jonasdottir A, Bjornsson A, Mattiasdottir S, Blondal T, Haraldsson M, Magnusdottir BB, Giegling I, Möller HJ, Hartmann A. et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium ISR. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi YY, He G, Zhang Z, Tang W, Zhang J Jr, Zhao Q, Zhang J Sr, Li XW, Xi ZR, Fang C, Zhao XZ, Feng GY, He L. A study of rare structural variants in schizophrenia patients and normal controls from Chinese Han population. Mol Psychiatry. 2008;13:911–913. doi: 10.1038/mp.2008.69. [DOI] [PubMed] [Google Scholar]
- Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, Conroy J, Magalhaes TR, Correia C, Abrahams BS, Almeida J, Bacchelli E, Bader GD, Bailey AJ, Baird G, Battaglia A, Berney T, Bolshakova N, Bölte S, Bolton PF, Bourgeron T, Brennan S, Brian J, Bryson SE, Carson AR, Casallo G, Casey J, Chung BH, Cochrane L, Corsello C. et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson DF, Duan J, Oh S, Wang K, Sanders AR, Shi J, Zhang N, Mowry BJ, Olincy A, Amin F, Cloninger CR, Silverman JM, Buccola NG, Byerley WF, Black DW, Kendler KS, Freedman R, Dudbridge F, Pe'er I, Hakonarson H, Bergen SE, Fanous AH, Holmans PA, Gejman PV. Copy number variants in schizophrenia: confirmation of five previous findings and new evidence for 3q29 microdeletions and VIPR2 duplications. Am J Psychiatry. 2011;168:302–316. doi: 10.1176/appi.ajp.2010.10060876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacic V, McCarthy S, Malhotra D, Murray F, Chou HH, Peoples A, Makarov V, Yoon S, Bhandari A, Corominas R, Iakoucheva LM, Krastoshevsky O, Krause V, Larach-Walters V, Welsh DK, Craig D, Kelsoe JR, Gershon ES, Leal SM, Dell Aquila M, Morris DW, Gill M, Corvin A, Insel PA, McClellan J, King MC, Karayiorgou M, Levy DL, DeLisi LE, Sebat J. Duplications of the neuropeptide receptor gene VIPR2 confer significant risk for schizophrenia. Nature. 2011;471:499–503. doi: 10.1038/nature09884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, Shago M, Moessner R, Pinto D, Ren Y, Thiruvahindrapduram B, Fiebig A, Schreiber S, Friedman J, Ketelaars CE, Vos YJ, Ficicioglu C, Kirkpatrick S, Nicolson R, Sloman L, Summers A, Gibbons CA, Teebi A, Chitayat D, Weksberg R, Thompson A, Vardy C, Crosbie V, Luscombe S, Baatjes R. et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glessner JT, Wang K, Cai G, Korvatska O, Kim CE, Wood S, Zhang H, Estes A, Brune CW, Bradfield JP, Imielinski M, Frackelton EC, Reichert J, Crawford EL, Munson J, Sleiman PM, Chiavacci R, Annaiah K, Thomas K, Hou C, Glaberson W, Flory J, Otieno F, Garris M, Soorya L, Klei L, Piven J, Meyer KJ, Anagnostou E, Sakurai T. et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, Ercan-Sencicek AG, Hus V, Luo R, Murtha MT, Moreno-De-Luca D, Chu SH, Moreau MP, Gupta AR, Thomson SA, Mason CE, Bilguvar K, Celestino-Soper PB, Choi M, Crawford EL, Davis L, Wright NR, Dhodapkar RM, DiCola M, DiLullo NM, Fernandez TV, Fielding-Singh V, Fishman DO, Frahm S, Garagaloyan R, Goh GS, Kammela S, Klei L, Lowe JK. et al. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70:863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia J, Gai X, Xie HM, Perin JC, Geiger E, Glessner JT, D'arcy M, deBerardinis R, Frackelton E, Kim C, Lantieri F, Muganga BM, Wang L, Takeda T, Rappaport EF, Grant SF, Berrettini W, Devoto M, Shaikh TH, Hakonarson H, White PS. Rare structural variants found in attention-deficit hyperactivity disorder are preferentially associated with neurodevelopmental genes. Mol Psychiatry. 2010;15:637–646. doi: 10.1038/mp.2009.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams NM, Zaharieva I, Martin A, Langley K, Mantripragada K, Fossdal R, Stefansson H, Stefansson K, Magnusson P, Gudmundsson OO, Gustafsson O, Holmans P, Owen MJ, O'Donovan M, Thapar A. Rare chromosomal deletions and duplications in attention-deficit hyperactivity disorder: a genome-wide analysis. Lancet. 2010;376:1401–1408. doi: 10.1016/S0140-6736(10)61109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionel AC, Crosbie J, Barbosa N, Goodale T, Thiruvahindrapuram B, Rickaby J, Gazzellone M, Carson AR, Howe JL, Wang Z, Wei J, Stewart AF, Roberts R, McPherson R, Fiebig A, Franke A, Schreiber S, Zwaigenbaum L, Fernandez BA, Roberts W, Arnold PD, Szatmari P, Marshall CR, Schachar R, Scherer SW. Rare copy number variation discovery and cross-disorder comparisons identify risk genes for ADHD. Sci Transl Med. 2011;3:95ra75. doi: 10.1126/scitranslmed.3002464. [DOI] [PubMed] [Google Scholar]
- Lesch KP, Selch S, Renner TJ, Jacob C, Nguyen TT, Hahn T, Romanos M, Walitza S, Shoichet S, Dempfle A, Heine M, Boreatti-Hümmer A, Romanos J, Gross-Lesch S, Zerlaut H, Wultsch T, Heinzel S, Fassnacht M, Fallgatter A, Allolio B, Schäfer H, Warnke A, Reif A, Ropers HH, Ullmann R. Genome-wide copy number variation analysis in attention-deficit/hyperactivity disorder: association with neuropeptide Y gene dosage in an extended pedigree. Mol Psychiatry. 2011;16:491–503. doi: 10.1038/mp.2010.29. [DOI] [PubMed] [Google Scholar]
- Sundaram SK, Huq AM, Wilson BJ, Chugani HT. Tourette syndrome is associated with recurrent exonic copy number variants. Neurology. 2010;74:1583–1590. doi: 10.1212/WNL.0b013e3181e0f147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, Carter NP, Church DM, Crolla JA, Eichler EE, Epstein CJ, Faucett WA, Feuk L, Friedman JM, Hamosh A, Jackson L, Kaminsky EB, Kok K, Krantz ID, Kuhn RM, Lee C, Ostell JM, Rosenberg C, Scherer SW, Spinner NB, Stavropoulos DJ, Tepperberg JH, Thorland EC, Vermeesch JR, Waggoner DJ, Watson MS. et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86:749–764. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper GM, Coe BP, Girirajan S, Rosenfeld JA, Vu TH, Baker C, Williams C, Stalker H, Hamid R, Hannig V, Abdel-Hamid H, Bader P, McCracken E, Niyazov D, Leppig K, Thiese H, Hummel M, Alexander N, Gorski J, Kussmann J, Shashi V, Johnson K, Rehder C, Ballif BC, Shaffer LG, Eichler EE. A copy number variation morbidity map of developmental delay. Nat Genet. 2011;43:838–846. doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mefford HC, Muhle H, Ostertag P, von Spiczak S, Buysse K, Baker C, Franke A, Malafosse A, Genton P, Thomas P, Gurnett CA, Schreiber S, Bassuk AG, Guipponi M, Stephani U, Helbig I, Eichler EE. Genome-wide copy number variation in epilepsy: novel susceptibility loci in idiopathic generalized and focal epilepsies. PLoS Genet. 2010;6:e1000962. doi: 10.1371/journal.pgen.1000962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariminejad R, Lind-Thomsen A, Tumer Z, Erdogan F, Ropers HH, Tommerup N, Ullmann R, Moller RS. High frequency of rare copy number variants affecting functionally related genes in patients with structural brain malformations. Hum Mutat. 2011. in press . [DOI] [PubMed]
- Piton A, Gauthier J, Hamdan FF, Lafrenière RG, Yang Y, Henrion E, Laurent S, Noreau A, Thibodeau P, Karemera L, Spiegelman D, Kuku F, Duguay J, Destroismaisons L, Jolivet P, Côté M, Lachapelle K, Diallo O, Raymond A, Marineau C, Champagne N, Xiong L, Gaspar C, Rivière JB, Tarabeux J, Cossette P, Krebs MO, Rapoport JL, Addington A, Delisi LE. et al. Systematic resequencing of X-chromosome synaptic genes in autism spectrum disorder and schizophrenia. Mol Psychiatry. 2010;16:867–880. doi: 10.1038/mp.2010.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissers LE, de Ligt J, Gilissen C, Janssen I, Steehouwer M, de Vries P, van Lier B, Arts P, Wieskamp N, del Rosario M, van Bon BW, Hoischen A, de Vries BB, Brunner HG, Veltman JA. A de novo paradigm for mental retardation. Nat Genet. 2010;42:1109–1112. doi: 10.1038/ng.712. [DOI] [PubMed] [Google Scholar]
- Glessner JT, Reilly MP, Kim CE, Takahashi N, Albano A, Hou C, Bradfield JP, Zhang H, Sleiman PM, Flory JH, Imielinski M, Frackelton EC, Chiavacci R, Thomas KA, Garris M, Otieno FG, Davidson M, Weiser M, Reichenberg A, Davis KL, Friedman JI, Cappola TP, Margulies KB, Rader DJ, Grant SF, Buxbaum JD, Gur RE, Hakonarson H. Strong synaptic transmission impact by copy number variations in schizophrenia. Proc Natl Acad Sci USA. 2010;107:10584–10589. doi: 10.1073/pnas.1000274107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Roos JL, Dexheimer P, Boone B, Plummer B, Levy S, Gogos JA, Karayiorgou M. Exome sequencing supports a de novo mutational paradigm for schizophrenia. Nat Genet. 2011;43:864–868. doi: 10.1038/ng.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard SL, Gauthier J, Noreau A, Xiong L, Zhou S, Jouan L, Dionne-Laporte A, Spiegelman D, Henrion E, Diallo O, Thibodeau P, Bachand I, Bao JY, Tong AH, Lin CH, Millet B, Jaafari N, Joober R, Dion PA, Lok S, Krebs MO, Rouleau GA. Increased exonic de novo mutation rate in individuals with schizophrenia. Nat Genet. 2011;43:860–863. doi: 10.1038/ng.886. [DOI] [PubMed] [Google Scholar]
- Myers RA, Casals F, Gauthier J, Hamdan FF, Keebler J, Boyko AR, Bustamante CD, Piton AM, Spiegelman D, Henrion E, Zilversmit M, Hussin J, Quinlan J, Yang Y, Lafrenière RG, Griffing AR, Stone EA, Rouleau GA, Awadalla P. A population genetic approach to mapping neurological disorder genes using deep resequencing. PLoS Genet. 2011;7:e1001318. doi: 10.1371/journal.pgen.1001318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Roak BJ, Deriziotis P, Lee C, Vives L, Schwartz JJ, Girirajan S, Karakoc E, Mackenzie AP, Ng SB, Baker C, Rieder MJ, Nickerson DA, Bernier R, Fisher SE, Shendure J, Eichler EE. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat Genet. 2011;43:585–589. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najmabadi H, Hu H, Garshasbi M, Zemojtel T, Abedini SS, Chen W, Hosseini M, Behjati F, Haas S, Jamali P, Zecha A, Mohseni M, Püttmann L, Vahid LN, Jensen C, Moheb LA, Bienek M, Larti F, Mueller I, Weissmann R, Darvish H, Wrogemann K, Hadavi V, Lipkowitz B, Esmaeeli-Nieh S, Wieczorek D, Kariminejad R, Firouzabadi SG, Cohen M, Fattahi Z. et al. Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature. 2011;478:57–63. doi: 10.1038/nature10423. [DOI] [PubMed] [Google Scholar]
- Friedman JI, Vrijenhoek T, Markx S, Janssen IM, van der Vliet WA, Faas BH, Knoers NV, Cahn W, Kahn RS, Edelmann L, Davis KL, Silverman JM, Brunner HG, van Kessel AG, Wijmenga C, Ophoff RA, Veltman JA. CNTNAP2 gene dosage variation is associated with schizophrenia and epilepsy. Mol Psychiatry. 2008;13:261–266. doi: 10.1038/sj.mp.4002049. [DOI] [PubMed] [Google Scholar]
- Mefford HC, Sharp AJ, Baker C, Itsara A, Jiang Z, Buysse K, Huang S, Maloney VK, Crolla JA, Baralle D, Collins A, Mercer C, Norga K, de Ravel T, Devriendt K, Bongers EM, de Leeuw N, Reardon W, Gimelli S, Bena F, Hennekam RC, Male A, Gaunt L, Clayton-Smith J, Simonic I, Park SM, Mehta SG, Nik-Zainal S, Woods CG, Firth HV. et al. Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. N Engl J Med. 2008;359:1685–1699. doi: 10.1056/NEJMoa0805384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilmatre A, Dubourg C, Mosca AL, Legallic S, Goldenberg A, Drouin-Garraud V, Layet V, Rosier A, Briault S, Bonnet-Brilhault F, Laumonnier F, Odent S, Le Vacon G, Joly-Helas G, David V, Bendavid C, Pinoit JM, Henry C, Impallomeni C, Germano E, Tortorella G, Di Rosa G, Barthelemy C, Andres C, Faivre L, Frébourg T, Saugier Veber P, Campion D. Recurrent rearrangements in synaptic and neurodevelopmental genes and shared biologic pathways in schizophrenia, autism, and mental retardation. Arch Gen Psychiatry. 2009;66:947–956. doi: 10.1001/archgenpsychiatry.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DT, Shen Y, Weiss LA, Korn J, Anselm I, Bridgemohan C, Cox GF, Dickinson H, Gentile J, Harris DJ, Hegde V, Hundley R, Khwaja O, Kothare S, Luedke C, Nasir R, Poduri A, Prasad K, Raffalli P, Reinhard A, Smith SE, Sobeih MM, Soul JS, Stoler J, Takeoka M, Tan WH, Thakuria J, Wolff R, Yusupov R, Gusella JF. et al. Microdeletion/duplication at 15q13.2q13.3 among individuals with features of autism and other neuropsychiatric disorders. J Med Genet. 2009;46:242–248. doi: 10.1136/jmg.2008.059907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ching MS, Shen Y, Tan WH, Jeste SS, Morrow EM, Chen X, Mukaddes NM, Yoo SY, Hanson E, Hundley R, Austin C, Becker RE, Berry GT, Driscoll K, Engle EC, Friedman S, Gusella JF, Hisama FM, Irons MB, Lafiosca T, LeClair E, Miller DT, Neessen M, Picker JD, Rappaport L, Rooney CM, Sarco DP, Stoler JM, Walsh CA, Wolff RR. et al. Deletions of NRXN1 (neurexin-1) predispose to a wide spectrum of developmental disorders. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:937–947. doi: 10.1002/ajmg.b.31063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell KJ. The genetics of neurodevelopmental disease. Curr Opin Neurobiol. 2011;21:197–203. doi: 10.1016/j.conb.2010.08.009. [DOI] [PubMed] [Google Scholar]
- Daniels JL, Forssen U, Hultman CM, Cnattingius S, Savitz DA, Feychting M, Sparen P. Parental psychiatric disorders associated with autism spectrum disorders in the offspring. Pediatrics. 2008;121:e1357–1362. doi: 10.1542/peds.2007-2296. [DOI] [PubMed] [Google Scholar]
- Lichtenstein P, Yip BH, Bjork C, Pawitan Y, Cannon TD, Sullivan PF, Hultman CM. Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: a population-based study. Lancet. 2009;373:234–239. doi: 10.1016/S0140-6736(09)60072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhausen HC, Foldager L, Perto G, Munk-Jorgensen P. Family aggregation of mental disorders in the nationwide Danish three generation study. Eur Arch Psychiatry Clin Neurosci. 2009;259:270–277. doi: 10.1007/s00406-008-0865-0. [DOI] [PubMed] [Google Scholar]
- Lichtenstein P, Carlstrom E, Rastam M, Gillberg C, Anckarsater H. The genetics of autism spectrum disorders and related neuropsychiatric disorders in childhood. Am J Psychiatry. 2010;167:1357–1363. doi: 10.1176/appi.ajp.2010.10020223. [DOI] [PubMed] [Google Scholar]
- Gottesman II, Laursen TM, Bertelsen A, Mortensen PB. Severe mental disorders in offspring with 2 psychiatrically ill parents. Arch Gen Psychiatry. 2010;67:252–257. doi: 10.1001/archgenpsychiatry.2010.1. [DOI] [PubMed] [Google Scholar]
- Chang Y-T, Chen P-C, Tsai I-J, Sung F-C, Chin Z-N, Kuo H-T, Tsai C-H, Chou I-C. Bidirectional relation between schizophrenia and epilepsy: A population-based retrospective cohort study. Epilepsia. 2011;52:2036–2042. doi: 10.1111/j.1528-1167.2011.03268.x. [DOI] [PubMed] [Google Scholar]
- Bassett AS, Scherer SW, Brzustowicz LM. Copy number variations in schizophrenia: critical review and new perspectives on concepts of genetics and disease. Am J Psychiatry. 2010;167:899–914. doi: 10.1176/appi.ajp.2009.09071016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chubb JE, Bradshaw NJ, Soares DC, Porteous DJ, Millar JK. The DISC locus in psychiatric illness. Mol Psychiatry. 2008;13:36–64. doi: 10.1038/sj.mp.4002106. [DOI] [PubMed] [Google Scholar]
- Raychaudhuri S, Plenge RM, Rossin EJ, Ng AC, Purcell SM, Sklar P, Scolnick EM, Xavier RJ, Altshuler D, Daly MJ. Identifying relationships among genomic disease regions: predicting genes at pathogenic SNP associations and rare deletions. PLoS Genet. 2009;5:e1000534. doi: 10.1371/journal.pgen.1000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancur C, Sakurai T, Buxbaum JD. The emerging role of synaptic cell-adhesion pathways in the pathogenesis of autism spectrum disorders. Trends Neurosci. 2009;32:402–412. doi: 10.1016/j.tins.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi-Takagi A, Sawa A. Disturbed synaptic connectivity in schizophrenia: convergence of genetic risk factors during neurodevelopment. Brain Res Bull. 2010;83:140–146. doi: 10.1016/j.brainresbull.2010.04.007. [DOI] [PubMed] [Google Scholar]
- Gilman SR, Iossifov I, Levy D, Ronemus M, Wigler M, Vitkup D. Rare de novo variants associated with autism implicate a large functional network of genes involved in formation and function of synapses. Neuron. 2011;70:898–907. doi: 10.1016/j.neuron.2011.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaha FJ, Quinlan JG. Dystrophinopathies: clarification and complication. J Child Neurol. 1996;11:13–20. doi: 10.1177/088307389601100103. [DOI] [PubMed] [Google Scholar]
- Blake DJ, Forrest M, Chapman RM, Tinsley CL, O'Donovan MC, Owen MJ. TCF4, schizophrenia, and Pitt-Hopkins syndrome. Schizophr Bull. 2010;36:443–447. doi: 10.1093/schbul/sbq035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow EM, Yoo SY, Flavell SW, Kim TK, Lin Y, Hill RS, Mukaddes NM, Balkhy S, Gascon G, Hashmi A, Al-Saad S, Ware J, Joseph RM, Greenblatt R, Gleason D, Ertelt JA, Apse KA, Bodell A, Partlow JN, Barry B, Yao H, Markianos K, Ferland RJ, Greenberg ME, Walsh CA. Identifying autism loci and genes by tracing recent shared ancestry. Science. 2008;321:218–223. doi: 10.1126/science.1157657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney JA. Genetic modifiers of neurological disease. Curr Opin Genet Dev. 2011;21:349–353. doi: 10.1016/j.gde.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girirajan S, Rosenfeld JA, Cooper GM, Antonacci F, Siswara P, Itsara A, Vives L, Walsh T, McCarthy SE, Baker C, Mefford HC, Kidd JM, Browning SR, Browning BL, Dickel DE, Levy DL, Ballif BC, Platky K, Farber DM, Gowans GC, Wetherbee JJ, Asamoah A, Weaver DD, Mark PR, Dickerson J, Garg BP, Ellingwood SA, Smith R, Banks VC, Smith W. et al. A recurrent 16p12.1 microdeletion supports a two-hit model for severe developmental delay. Nat Genet. 2010;42:203–209. doi: 10.1038/ng.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klassen T, Davis C, Goldman A, Burgess D, Chen T, Wheeler D, McPherson J, Bourquin T, Lewis L, Villasana D, Morgan M, Muzny D, Gibbs R, Noebels J. Exome sequencing of ion channel genes reveals complex profiles confounding personal risk assessment in epilepsy. Cell. 2011;145:1036–1048. doi: 10.1016/j.cell.2011.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaf CP, Sabo A, Sakai Y, Crosby J, Muzny D, Hawes A, Lewis L, Akbar H, Varghese R, Boerwinkle E, Gibbs RA, Zoghbi HY. Oligogenic heterozygosity in individuals with high-functioning autism spectrum disorders. Hum Mol Genet. 2011;20:3366–3375. doi: 10.1093/hmg/ddr243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell KJ. The genetics of brain wiring: from molecule to mind. PLoS Biol. 2007;5:e113. doi: 10.1371/journal.pbio.0050113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tandon R, Keshavan MS, Nasrallah HA. Schizophrenia, "just the facts" what we know in 2008. 2. Epidemiology and etiology. Schizophr Res. 2008;102:1–18. doi: 10.1016/j.schres.2008.04.011. [DOI] [PubMed] [Google Scholar]
- van der Weyden L, Bradley A. Mouse chromosome engineering for modeling human disease. Annu Rev Genomics Hum Genet. 2006;7:247–276. doi: 10.1146/annurev.genom.7.080505.115741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, Mujica AO, Thomas M, Harrow J, Cox T, Jackson D, Severin J, Biggs P, Fu J, Nefedov M, de Jong PJ, Stewart AF, Bradley A. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi K, Blundell J, Etherton MR, Hammer RE, Liu X, Powell CM, Sudhof TC. A neuroligin-3 mutation implicated in autism increases inhibitory synaptic transmission in mice. Science. 2007;318:71–76. doi: 10.1126/science.1146221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etherton M, Foldy C, Sharma M, Tabuchi K, Liu X, Shamloo M, Malenka RC, Sudhof TC. Autism-linked neuroligin-3 R451C mutation differentially alters hippocampal and cortical synaptic function. Proc Natl Acad Sci USA. 2011;108:13764–13769. doi: 10.1073/pnas.1111093108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangash MA, Park JM, Melnikova T, Wang D, Jeon SK, Lee D, Syeda S, Kim J, Kouser M, Schwartz J, Cui Y, Zhao X, Speed HE, Kee SE, Tu JC, Hu JH, Petralia RS, Linden DJ, Powell CM, Savonenko A, Xiao B, Worley PF. Enhanced polyubiquitination of Shank3 and NMDA receptor in a mouse model of autism. Cell. 2011;145:758–772. doi: 10.1016/j.cell.2011.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Nakatani J, Tamada K, Hatanaka F, Ise S, Ohta H, Inoue K, Tomonaga S, Watanabe Y, Chung YJ, Banerjee R, Iwamoto K, Kato T, Okazawa M, Yamauchi K, Tanda K, Takao K, Miyakawa T, Bradley A, Takumi T. Abnormal behavior in a chromosome-engineered mouse model for human 15q11-13 duplication seen in autism. Cell. 2009;137:1235–1246. doi: 10.1016/j.cell.2009.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurdsson T, Stark KL, Karayiorgou M, Gogos JA, Gordon JA. Impaired hippocampal-prefrontal synchrony in a genetic mouse model of schizophrenia. Nature. 2010;464:763–767. doi: 10.1038/nature08855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horev G, Ellegood J, Lerch JP, Son YE, Muthuswamy L, Vogel H, Krieger AM, Buja A, Henkelman RM, Wigler M, Mills AA. Dosage-dependent phenotypes in models of 16p11.2 lesions found in autism. Proc Natl Acad Sci USA. 2011;108:17076–17081. doi: 10.1073/pnas.1114042108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karayiorgou M, Simon TJ, Gogos JA. 22q11.2 microdeletions: linking DNA structural variation to brain dysfunction and schizophrenia. Nat Rev Neurosci. 2010;11:402–416. doi: 10.1038/nrn2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SE, Zhou YD, Zhang G, Jin Z, Stoppel DC, Anderson MP. Increased gene dosage of ube3a results in autism traits and decreased glutamate synaptic transmission in mice. Sci Transl Med. 2011;3:103ra197. doi: 10.1126/scitranslmed.3002627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etherton MR, Blaiss CA, Powell CM, Sudhof TC. Mouse neurexin-1alpha deletion causes correlated electrophysiological and behavioral changes consistent with cognitive impairments. Proc Natl Acad Sci USA. 2009;106:17998–18003. doi: 10.1073/pnas.0910297106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozdagi O, Sakurai T, Papapetrou D, Wang X, Dickstein DL, Takahashi N, Kajiwara Y, Yang M, Katz AM, Scattoni ML, Harris MJ, Saxena R, Silverman JL, Crawley JN, Zhou Q, Hof PR, Buxbaum JD. Haploinsufficiency of the autism-associated Shank3 gene leads to deficits in synaptic function, social interaction, and social communication. Mol Autism. 2010;1:15. doi: 10.1186/2040-2392-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peca J, Feliciano C, Ting JT, Wang W, Wells MF, Venkatraman TN, Lascola CD, Fu Z, Feng G. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature. 2011;472:437–442. doi: 10.1038/nature09965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peñagarikano O, Abrahams BS, Herman EI, Winden KD, Gdalyahu A, Dong H, Sonnenblick LI, Gruver R, Almajano J, Bragin A, Golshani P, Trachtenberg JT, Peles E, Geschwind DH. Absence of CNTNAP2 leads to epilepsy, neuronal migration abnormalities, and core autism-related deficits. Cell. 2011;147:235–246. doi: 10.1016/j.cell.2011.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvajo M, McKellar H, Arguello PA, Drew LJ, Moore H, MacDermott AB, Karayiorgou M, Gogos JA. A mutation in mouse Disc1 that models a schizophrenia risk allele leads to specific alterations in neuronal architecture and cognition. Proc Natl Acad Sci USA. 2008;105:7076–7081. doi: 10.1073/pnas.0802615105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee FH, Fadel MP, Preston-Maher K, Cordes SP, Clapcote SJ, Price DJ, Roder JC, Wong AH. Disc1 point mutations in mice affect development of the cerebral cortex. J Neurosci. 2011;31:3197–3206. doi: 10.1523/JNEUROSCI.4219-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osbun N, Li J, O'Driscoll MC, Strominger Z, Wakahiro M, Rider E, Bukshpun P, Boland E, Spurrell CH, Schackwitz W, Pennacchio LA, Dobyns WB, Black GC, Sherr EH. Genetic and functional analyses identify DISC1 as a novel callosal agenesis candidate gene. Am J Med Genet A. 2011;155A:1865–1876. doi: 10.1002/ajmg.a.34081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohal VS, Zhang F, Yizhar O, Deisseroth K. Parvalbumin neurons and gamma rhythms enhance cortical circuit performance. Nature. 2009;459:698–702. doi: 10.1038/nature07991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yizhar O, Fenno LE, Davidson TJ, Mogri M, Deisseroth K. Optogenetics in neural systems. Neuron. 2011;71:9–34. doi: 10.1016/j.neuron.2011.06.004. [DOI] [PubMed] [Google Scholar]
- Yizhar O, Fenno LE, Prigge M, Schneider F, Davidson TJ, O'Shea DJ, Sohal VS, Goshen I, Finkelstein J, Paz JT, Stehfest K, Fudim R, Ramakrishnan C, Huguenard JR, Hegemann P, Deisseroth K. Neocortical excitation/inhibition balance in information processing and social dysfunction. Nature. 2011;477:171–178. doi: 10.1038/nature10360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolen G, Osterweil E, Rao BS, Smith GB, Auerbach BD, Chattarji S, Bear MF. Correction of fragile × syndrome in mice. Neuron. 2007;56:955–962. doi: 10.1016/j.neuron.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehninger D, Li W, Fox K, Stryker MP, Silva AJ. Reversing neurodevelopmental disorders in adults. Neuron. 2008;60:950–960. doi: 10.1016/j.neuron.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tropea D, Giacometti E, Wilson NR, Beard C, McCurry C, Fu DD, Flannery R, Jaenisch R, Sur M. Partial reversal of Rett Syndrome-like symptoms in MeCP2 mutant mice. Proc Natl Acad Sci USA. 2009;106:2029–2034. doi: 10.1073/pnas.0812394106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hays SA, Huber KM, Gibson JR. Altered neocortical rhythmic activity states in Fmr1 KO mice are due to enhanced mGluR5 signaling and involve changes in excitatory circuitry. J Neurosci. 2011;31:14223–14234. doi: 10.1523/JNEUROSCI.3157-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chubykin AA, Atasoy D, Etherton MR, Brose N, Kavalali ET, Gibson JR, Sudhof TC. Activity-dependent validation of excitatory versus inhibitory synapses by neuroligin-1 versus neuroligin-2. Neuron. 2007;54:919–931. doi: 10.1016/j.neuron.2007.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blundell J, Blaiss CA, Etherton MR, Espinosa F, Tabuchi K, Walz C, Bolliger MF, Sudhof TC, Powell CM. Neuroligin-1 deletion results in impaired spatial memory and increased repetitive behavior. J Neurosci. 2010;30:2115–2129. doi: 10.1523/JNEUROSCI.4517-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedlicka P, Hoon M, Papadopoulos T, Vlachos A, Winkels R, Poulopoulos A, Betz H, Deller T, Brose N, Varoqueaux F, Schwarzacher SW. Increased dentate gyrus excitability in neuroligin-2-deficient mice in vivo. Cereb Cortex. 2011;21:357–367. doi: 10.1093/cercor/bhq100. [DOI] [PubMed] [Google Scholar]
- Gibson JR, Huber KM, Sudhof TC. Neuroligin-2 deletion selectively decreases inhibitory synaptic transmission originating from fast-spiking but not from somatostatin-positive interneurons. J Neurosci. 2009;29:13883–13897. doi: 10.1523/JNEUROSCI.2457-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazzari P, Paternain AV, Valiente M, Pla R, Lujan R, Lloyd K, Lerma J, Marin O, Rico B. Control of cortical GABA circuitry development by Nrg1 and ErbB4 signalling. Nature. 2010;464:1376–1380. doi: 10.1038/nature08928. [DOI] [PubMed] [Google Scholar]
- Lisman JE, Coyle JT, Green RW, Javitt DC, Benes FM, Heckers S, Grace AA. Circuit-based framework for understanding neurotransmitter and risk gene interactions in schizophrenia. Trends Neurosci. 2008;31:234–242. doi: 10.1016/j.tins.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howes OD, Kapur S. The dopamine hypothesis of schizophrenia: version III--the final common pathway. Schizophr Bull. 2009;35:549–562. doi: 10.1093/schbul/sbp006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grace AA. Dopamine system dysregulation by the ventral subiculum as the common pathophysiological basis for schizophrenia psychosis, psychostimulant abuse, and stress. Neurotox Res. 2010;18:367–376. doi: 10.1007/s12640-010-9154-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Burgos G, Fish KN, Lewis DA. GABA neuron alterations, cortical circuit dysfunction and cognitive deficits in schizophrenia. Neural Plast. 2011;2011:723184. doi: 10.1155/2011/723184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Callaway EM, Svoboda K. Genetic dissection of neural circuits. Neuron. 2008;57:634–660. doi: 10.1016/j.neuron.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenkiel BR, Ehlers MD. Molecular genetics and imaging technologies for circuit-based neuroanatomy. Nature. 2009;461:900–907. doi: 10.1038/nature08536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohland JW, Wu C, Barbas H, Bokil H, Bota M, Breiter HC, Cline HT, Doyle JC, Freed PJ, Greenspan RJ, Haber SN, Hawrylycz M, Herrera DG, Hilgetag CC, Huang ZJ, Jones A, Jones EG, Karten HJ, Kleinfeld D, Kötter R, Lester HA, Lin JM, Mensh BD, Mikula S, Panksepp J, Price JL, Safdieh J, Saper CB, Schiff ND, Schmahmann JD. et al. A proposal for a coordinated effort for the determination of brainwide neuroanatomical connectivity in model organisms at a mesoscopic scale. PLoS Comput Biol. 2009;5:e1000334. doi: 10.1371/journal.pcbi.1000334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor DH, Huber D, Svoboda K. Reverse engineering the mouse brain. Nature. 2009;461:923–929. doi: 10.1038/nature08539. [DOI] [PubMed] [Google Scholar]
- Morozov A. Conditional gene expression and targeting in neuroscience research. Curr Protoc Neurosci. 2008;Chapter 4 doi: 10.1002/0471142301.ns0431s44. Unit 4 31. [DOI] [PubMed] [Google Scholar]
- Gong S, Doughty M, Harbaugh CR, Cummins A, Hatten ME, Heintz N, Gerfen CR. Targeting Cre recombinase to specific neuron populations with bacterial artificial chromosome constructs. J Neurosci. 2007;27:9817–9823. doi: 10.1523/JNEUROSCI.2707-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi H, He M, Wu P, Kim S, Paik R, Sugino K, Kvitsani D, Fu Y, Lu J, Lin Y, Miyoshi G, Shima Y, Fishell G, Nelson SB, Huang ZJ. A resource of cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron. 2011;71:995–1013. doi: 10.1016/j.neuron.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar JK, Pickard BS, Mackie S, James R, Christie S, Buchanan SR, Malloy MP, Chubb JE, Huston E, Baillie GS, Thomson PA, Hill EV, Brandon NJ, Rain JC, Camargo LM, Whiting PJ, Houslay MD, Blackwood DH, Muir WJ, Porteous DJ. DISC1 and PDE4B are interacting genetic factors in schizophrenia that regulate cAMP signaling. Science. 2005;310:1187–1191. doi: 10.1126/science.1112915. [DOI] [PubMed] [Google Scholar]
- Seeman P. All roads to schizophrenia lead to dopamine supersensitivity and elevated dopamine D2(high) receptors. CNS Neurosci Ther. 2011;17:118–132. doi: 10.1111/j.1755-5949.2010.00162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden C. Parsing the genetics of behavior. Science. 2008;322:892–895. doi: 10.1126/science.322.5903.892. [DOI] [PubMed] [Google Scholar]
- Singer W. Development and plasticity of cortical processing architectures. Science. 1995;270:758–764. doi: 10.1126/science.270.5237.758. [DOI] [PubMed] [Google Scholar]
- Williams MA, Sachdev PS. Magnetoencephalography in neuropsychiatry: ready for application? Curr Opin Psychiatry. 2010;23:273–277. doi: 10.1097/YCO.0b013e328338621d. [DOI] [PubMed] [Google Scholar]
- Fries P, Reynolds JH, Rorie AE, Desimone R. Modulation of oscillatory neuronal synchronization by selective visual attention. Science. 2001;291:1560–1563. doi: 10.1126/science.1055465. [DOI] [PubMed] [Google Scholar]
- Buzsaki G. Large-scale recording of neuronal ensembles. Nat Neurosci. 2004;7:446–451. doi: 10.1038/nn1233. [DOI] [PubMed] [Google Scholar]
- Uhlhaas PJ, Roux F, Rodriguez E, Rotarska-Jagiela A, Singer W. Neural synchrony and the development of cortical networks. Trends Cogn Sci. 2010;14:72–80. doi: 10.1016/j.tics.2009.12.002. [DOI] [PubMed] [Google Scholar]
- Spencer KM, Nestor PG, Niznikiewicz MA, Salisbury DF, Shenton ME, McCarley RW. Abnormal neural synchrony in schizophrenia. J Neurosci. 2003;23:7407–7411. doi: 10.1523/JNEUROSCI.23-19-07407.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlhaas PJ, Linden DE, Singer W, Haenschel C, Lindner M, Maurer K, Rodriguez E. Dysfunctional long-range coordination of neural activity during Gestalt perception in schizophrenia. J Neurosci. 2006;26:8168–8175. doi: 10.1523/JNEUROSCI.2002-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlhaas PJ, Singer W. Abnormal neural oscillations and synchrony in schizophrenia. Nat Rev Neurosci. 2010;11:100–113. doi: 10.1038/nrn2774. [DOI] [PubMed] [Google Scholar]
- Bob P, Susta M, Glaslova K, Boutros NN. Dissociative symptoms and interregional EEG cross-correlations in paranoid schizophrenia. Psychiatry Res. 2010;177:37–40. doi: 10.1016/j.psychres.2009.08.015. [DOI] [PubMed] [Google Scholar]
- Belforte JE, Zsiros V, Sklar ER, Jiang Z, Yu G, Li Y, Quinlan EM, Nakazawa K. Postnatal NMDA receptor ablation in corticolimbic interneurons confers schizophrenia-like phenotypes. Nat Neurosci. 2010;13:76–83. doi: 10.1038/nn.2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakami T, Jones NC, Tolmacheva EA, Gaudias J, Chaumont J, Salzberg M, O'Brien TJ, Pinault D. NMDA receptor hypofunction leads to generalized and persistent aberrant gamma oscillations independent of hyperlocomotion and the state of consciousness. PLoS ONE. 2009;4:e6755. doi: 10.1371/journal.pone.0006755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirota A, Montgomery S, Fujisawa S, Isomura Y, Zugaro M, Buzsaki G. Entrainment of neocortical neurons and gamma oscillations by the hippocampal theta rhythm. Neuron. 2008;60:683–697. doi: 10.1016/j.neuron.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardin JA, Carlen M, Meletis K, Knoblich U, Zhang F, Deisseroth K, Tsai LH, Moore CI. Driving fast-spiking cells induces gamma rhythm and controls sensory responses. Nature. 2009;459:663–667. doi: 10.1038/nature08002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fries P. Neuronal gamma-band synchronization as a fundamental process in cortical computation. Annu Rev Neurosci. 2009;32:209–224. doi: 10.1146/annurev.neuro.051508.135603. [DOI] [PubMed] [Google Scholar]
- Korotkova T, Fuchs EC, Ponomarenko A, von Engelhardt J, Monyer H. NMDA receptor ablation on parvalbumin-positive interneurons impairs hippocampal synchrony, spatial representations, and working memory. Neuron. 2010;68:557–569. doi: 10.1016/j.neuron.2010.09.017. [DOI] [PubMed] [Google Scholar]
- Colgin LL. Oscillations and hippocampal-prefrontal synchrony. Curr Opin Neurobiol. 2011;21:467–474. doi: 10.1016/j.conb.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klausberger T, Marton LF, O'Neill J, Huck JH, Dalezios Y, Fuentealba P, Suen WY, Papp E, Kaneko T, Watanabe M, Csicsvari J, Somogyi P. Complementary roles of cholecystokinin- and parvalbumin-expressing GABAergic neurons in hippocampal network oscillations. J Neurosci. 2005;25:9782–9793. doi: 10.1523/JNEUROSCI.3269-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentealba P, Begum R, Capogna M, Jinno S, Marton LF, Csicsvari J, Thomson A, Somogyi P, Klausberger T. Ivy cells: a population of nitric-oxide-producing, slow-spiking GABAergic neurons and their involvement in hippocampal network activity. Neuron. 2008;57:917–929. doi: 10.1016/j.neuron.2008.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwich K, Pollak T, Klausberger T. Distinct firing patterns of identified basket and dendrite-targeting interneurons in the prefrontal cortex during hippocampal theta and local spindle oscillations. J Neurosci. 2009;29:9563–9574. doi: 10.1523/JNEUROSCI.1397-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacson JS, Scanziani M. How inhibition shapes cortical activity. Neuron. 2011;72:231–243. doi: 10.1016/j.neuron.2011.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tye KM, Prakash R, Kim SY, Fenno LE, Grosenick L, Zarabi H, Thompson KR, Gradinaru V, Ramakrishnan C, Deisseroth K. Amygdala circuitry mediating reversible and bidirectional control of anxiety. Nature. 2011;471:358–362. doi: 10.1038/nature09820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll D. Genome engineering with zinc-finger nucleases. Genetics. 2011;188:773–782. doi: 10.1534/genetics.111.131433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geurts AM, Cost GJ, Freyvert Y, Zeitler B, Miller JC, Choi VM, Jenkins SS, Wood A, Cui X, Meng X, Vincent A, Lam S, Michalkiewicz M, Schilling R, Foeckler J, Kalloway S, Weiler H, Ménoret S, Anegon I, Davis GD, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Jacob HJ, Buelow R. Knockout rats via embryo microinjection of zinc-finger nucleases. Science. 2009;325:433. doi: 10.1126/science.1172447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Ji D, Fisher DA, Wu Y, Briner DM, Weinstein EJ. Targeted integration in rat and mouse embryos with zinc-finger nucleases. Nat Biotechnol. 2011;29:64–67. doi: 10.1038/nbt.1731. [DOI] [PubMed] [Google Scholar]
- Huang G, Ashton C, Kumbhani DS, Ying QL. Genetic manipulations in the rat: progress and prospects. Curr Opin Nephrol Hypertens. 2011;20:391–399. doi: 10.1097/MNH.0b013e328347768a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betley JN, Sternson SM. Adeno-associated viral vectors for mapping, monitoring, and manipulating neural circuits. Hum Gene Ther. 2011;22:669–677. doi: 10.1089/hum.2010.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Qian X, Bernstein JG, Zhou HH, Franzesi GT, Stern P, Bronson RT, Graybiel AM, Desimone R, Boyden ES. Millisecond-timescale optical control of neural dynamics in the nonhuman primate brain. Neuron. 2009;62:191–198. doi: 10.1016/j.neuron.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdyyeva TK, Reynolds JH. The dawning of primate optogenetics. Neuron. 2009;62:159–160. doi: 10.1016/j.neuron.2009.04.011. [DOI] [PubMed] [Google Scholar]
- Carter CS, Barch DM. Cognitive neuroscience-based approaches to measuring and improving treatment effects on cognition in schizophrenia: the CNTRICS initiative. Schizophr Bull. 2007;33:1131–1137. doi: 10.1093/schbul/sbm081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Lindenberg A, Poline JB, Kohn PD, Holt JL, Egan MF, Weinberger DR, Berman KF. Evidence for abnormal cortical functional connectivity during working memory in schizophrenia. Am J Psychiatry. 2001;158:1809–1817. doi: 10.1176/appi.ajp.158.11.1809. [DOI] [PubMed] [Google Scholar]
- Bertolino A, Rubino V, Sambataro F, Blasi G, Latorre V, Fazio L, Caforio G, Petruzzella V, Kolachana B, Hariri A, Meyer-Lindenberg A, Nardini M, Weinberger DR, Scarabino T. Prefrontal-hippocampal coupling during memory processing is modulated by COMT val158met genotype. Biol Psychiatry. 2006;60:1250–1258. doi: 10.1016/j.biopsych.2006.03.078. [DOI] [PubMed] [Google Scholar]
- van den Heuvel MP, Hulshoff Pol HE. Exploring the brain network: a review on resting-state fMRI functional connectivity. Eur Neuropsychopharmacol. 2010;20:519–534. doi: 10.1016/j.euroneuro.2010.03.008. [DOI] [PubMed] [Google Scholar]
- Schipul SE, Keller TA, Just MA. Inter-regional brain communication and its disturbance in autism. Front Syst Neurosci. 2011;5:10. doi: 10.3389/fnsys.2011.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willi R, Weinmann O, Winter C, Klein J, Sohr R, Schnell L, Yee BK, Feldon J, Schwab ME. Constitutive genetic deletion of the growth regulator Nogo-A induces schizophrenia-related endophenotypes. J Neurosci. 2010;30:556–567. doi: 10.1523/JNEUROSCI.4393-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feyder M, Karlsson RM, Mathur P, Lyman M, Bock R, Momenan R, Munasinghe J, Scattoni ML, Ihne J, Camp M, Graybeal C, Strathdee D, Begg A, Alvarez VA, Kirsch P, Rietschel M, Cichon S, Walter H, Meyer-Lindenberg A, Grant SG, Holmes A. Association of mouse Dlg4 (PSD-95) gene deletion and human DLG4 gene variation with phenotypes relevant to autism spectrum disorders and Williams' syndrome. Am J Psychiatry. 2010;167:1508–1517. doi: 10.1176/appi.ajp.2010.10040484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rünker AE, O'Tuathaigh CM, Dunleavy M, Morris DW, Little GE, Corvin AP, Gill M, Henshall DC, Waddington JL, Mitchell KJ. Mutation of semaphorin-6A disrupts limbic and cortical connectivity and models neurodevelopmental psychopathology. PLoS ONE. 2011. in press . [DOI] [PMC free article] [PubMed]
- Shmelkov SV, Hormigo A, Jing D, Proenca CC, Bath KG, Milde T, Shmelkov E, Kushner JS, Baljevic M, Dincheva I, Murphy AJ, Valenzuela DM, Gale NW, Yancopoulos GD, Ninan I, Lee FS, Rafii S. Slitrk5 deficiency impairs corticostriatal circuitry and leads to obsessive-compulsive-like behaviors in mice. Nat Med. 2010;16:598–602. doi: 10.1038/nm.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tandon R, Belmaker RH, Gattaz WF, Lopez-Ibor JJ Jr, Okasha A, Singh B, Stein DJ, Olie JP, Fleischhacker WW, Moeller HJ. World Psychiatric Association Pharmacopsychiatry Section statement on comparative effectiveness of antipsychotics in the treatment of schizophrenia. Schizophr Res. 2008;100:20–38. doi: 10.1016/j.schres.2007.11.033. [DOI] [PubMed] [Google Scholar]
- Bryson SE, Rogers SJ, Fombonne E. Autism spectrum disorders: early detection, intervention, education, and psychopharmacological management. Can J Psychiatry. 2003;48:506–516. doi: 10.1177/070674370304800802. [DOI] [PubMed] [Google Scholar]
- Nagai T, Kitahara Y, Ibi D, Nabeshima T, Sawa A, Yamada K. Effects of antipsychotics on the behavioral deficits in human dominant-negative DISC1 transgenic mice with neonatal polyI:C treatment. Behav Brain Res. 2011;225:305–310. doi: 10.1016/j.bbr.2011.07.049. [DOI] [PubMed] [Google Scholar]