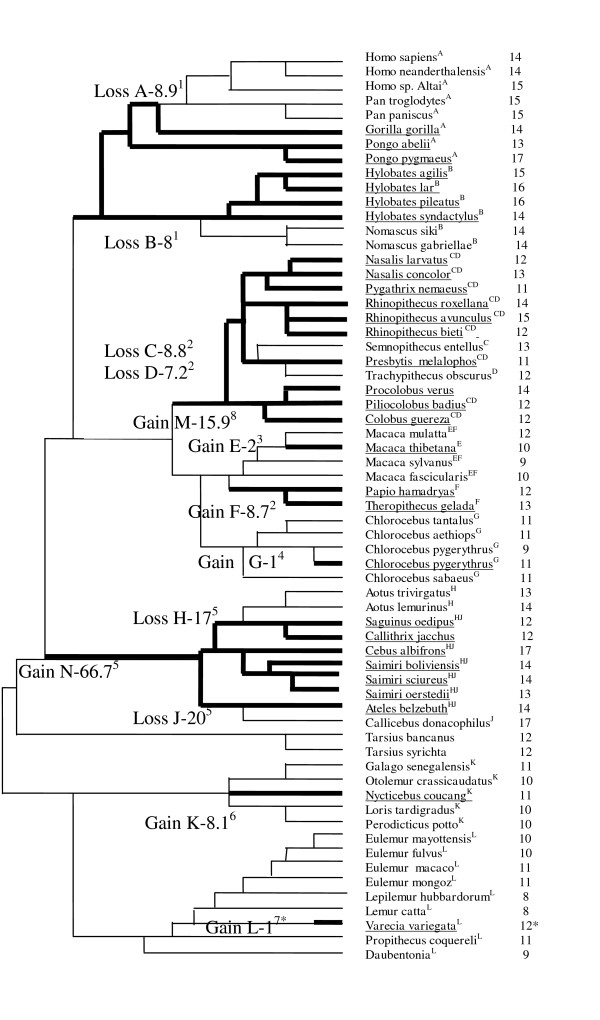
Figure 3.
Schematic representation of the phylogenetic relationship between the primate species examined for gau followed by numbers of stops in this region. Underlined species possess a predicted mitochondrial antisense antitermination AGR tRNA, which potentially enables translation of the AGR codons without importing cytosolic tRNAs. Broad branches represent evolution in the presence of antitermination tRNAs, as predicted by parsimony. Branch lengths are not proportional to divergence times, but specific nodes used for contrast analyses between lineages with and without antitermination AGR tRNA are indicated by capital letters, together with estimated divergence times, because these lineages with and without antitermination AGR tRNA diverged. Divergence times (millions of years) shown are from the following references (superscripts match the corresponding nodes indicated by capital letters): [27]1, [28]2, [29]3, [30]4, [31]5, [32]6, [33]7 and [34]8. *Analyses of several sequences indicate the possibility of a polymorphism in the presence of the antitermination AGR tRNA in Varecia. It is most parsimonious to assume that the presence of antitermination AGR tRNA is very recent in this genus.