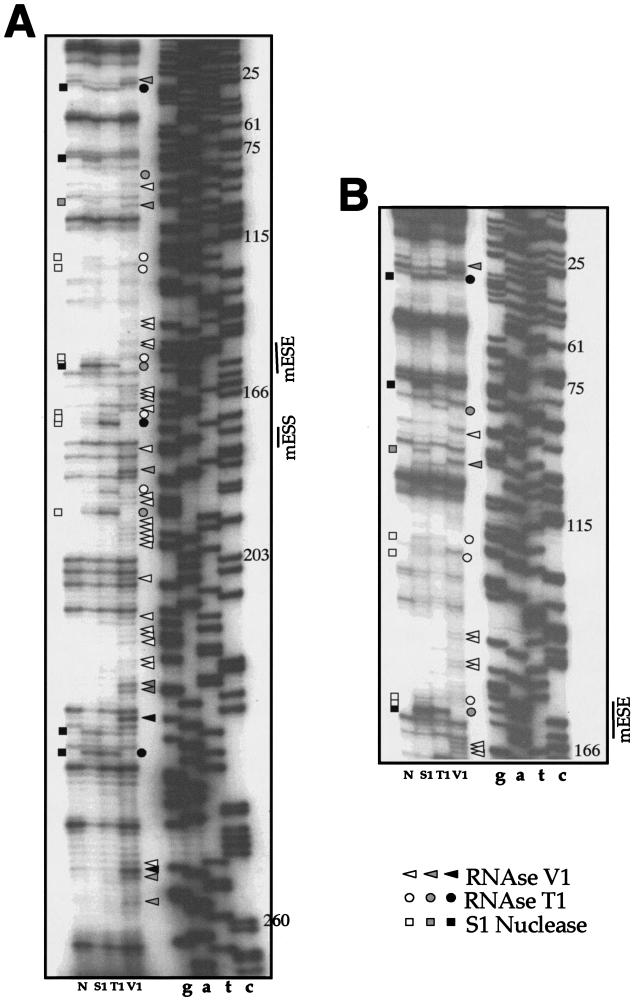
FIG.3.
Enzymatic determination of the RNA secondary structure of the mouse EDA exon. In vitro-transcribed mTot RNA was enzymatically digested with S1 nuclease and RNases T1 and V1 and reverse transcribed with an antisense primer. The RT products were separated on a sequencing polyacrylamide gel. A sequencing reaction (numbered according to the exon length) performed with the same primer was run in parallel with the cleavages in order to precisely determine the cleavage sites. In order to cover the whole exon length, short (A) and long (B) runs were performed. Squares, circles, and triangles indicate S1 nuclease and RNase T1 and V1 cleavage sites, respectively. Black, shaded, and white symbols indicate high, medium, and low cleavage intensities, respectively. No enzyme was added to the reaction mixture in lane N. The mESS and mESE positions are marked on the right of each sequencing reaction.