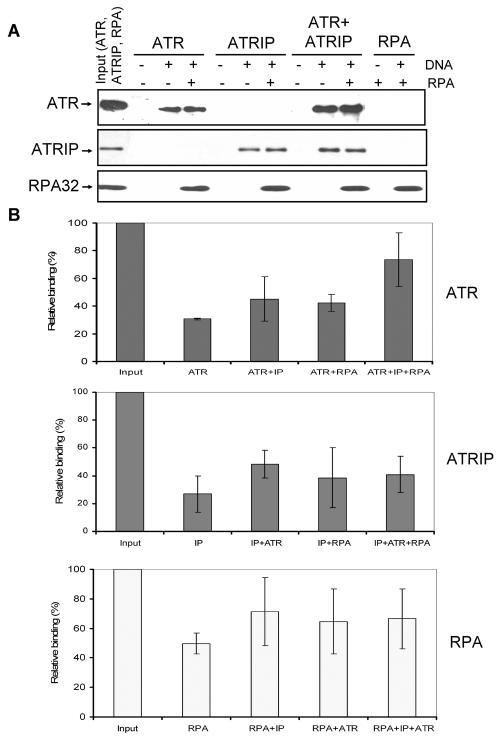
FIG. 6.
Effects of RPA and ATRIP on binding of ATR to DNA. (A) An 80-nt 5′-biotinylated ssDNA (7 pmol) was immobilized on streptavidin beads, and where indicated RPA was added to the beads and incubated for 30 min at 30°C before the addition of other proteins as shown. For each binding reaction, 4 pmol of ATR, 4 pmol of ATRIP, and 7 pmol of RPA were used. Following addition of proteins to the beads and 30 min of incubation at 30°C, the beads were extensively washed and the bound proteins were identified by Western blotting. Input represents one-half of the total protein used in the experiment. The sources of proteins used in the binding assays were as follows. ATR is Flag-ATR purified from transfected HEK293T cells by a high-salt wash of the affinity resin and lacks detectable ATRIP by Western analysis; ATRIP was purified from baculovirus-infected Sf21 cells; the ATR + ATRIP complex was obtained by mixing and preincubating the two proteins prior to adding to the binding reaction mixture. Note that even though we used a mixture of ATR plus ATRIP to obtain the ATR-ATRIP complex for our binding assays as was done previously (43), we obtained similar properties with the ATR-ATRIP complex copurified from cells cotransfected with vectors expressing both proteins (data not shown). RPA was purified from an E. coli strain expressing all three subunits of the protein. (B) Quantitative analysis of the DNA binding data for ATR (top), ATRIP (middle), and RPA (bottom) from three experiments carried out under conditions described for panel A. The bound protein is expressed in terms of percentage of the input, and the bars indicate standard errors. There was statistically significant (P < 0.05) difference only between the binding of ATR in isolation and the binding of ATR in the ATR + ATRIP complex to RPA-covered DNA (top panel, last two columns).