FIG. 4.
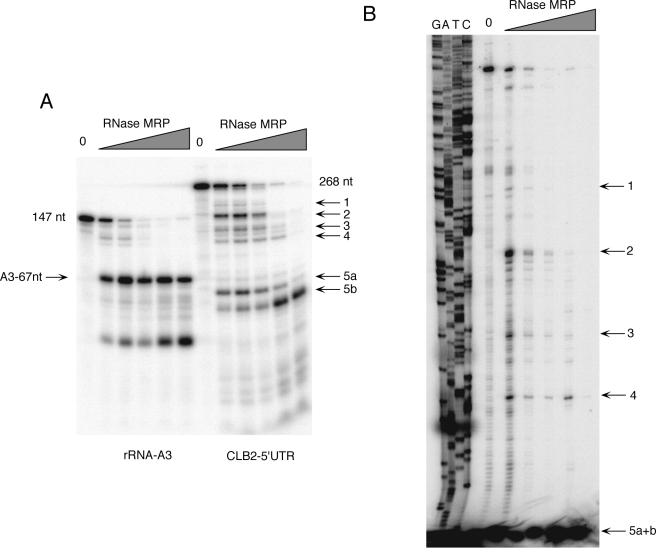
In vitro cleavage of the CLB2 5′-UTR and mapping of cleavage sites. (A) 3′-end-labeled RNA substrates containing the rRNA A3 cleavage site (positive control) and a 268-nucleotide CLB2 5′-UTR were generated. Highly pure yeast RNase MRP was purified with the TAP tag protocol and used in a standard RNase MRP in vitro assay (3, 22). Increasing amounts of enzyme were used in the assay: from left to right, 0 ng, 20 ng, 40 ng, 80 ng, 200 ng, and 800 ng of protein for both substrates. (B) A second RNase MRP assay was performed exactly as in panel A but then subjected to primer extension with the primer O-CLB2-12 (34). The positions of the cleavage sites are shown in Fig. 5.