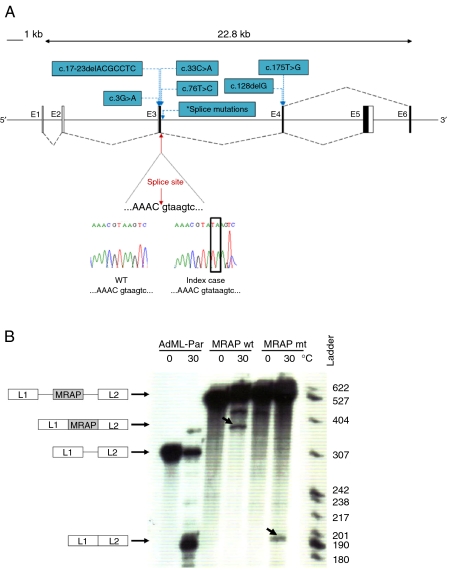
Figure 2.
(A) Schematic diagram representing alternative splicing of MRAP exons 5 and 6. Black boxes represent coding regions and black dashed lines indicate splicing. Chromatogram showing the splice mutation (red arrow). Other known MRAP mutations are shown in blue. *Indicates position of splice-site mutations (c.106+1G>T, c.106+1G>A, c.106+1G>C, c.106+1delG and c.106+2insT). (B) In vitro splicing assay demonstrates that MRAP mutation c.106+2_3dupTA leads to impaired splicing and skipping of exon 3. In this assay, MRAP exon 3 flanked by either wild type (wt) or mutated (mt; c.106+2_3dupTA) MRAP intronic sequences were introduced into a splicing reporter (AdML-Par) between the first and the second leader exons, L1 and L2. The resulting minigenes, MRAP wt (L1-MRAP wt_exon3-L2), MRAP mt (L1-MRAP mt_exon3-L2) or AdML-Par (splicing reporter alone, L1–L2), were incubated in HeLa nuclear extracts for 60 min under splicing conditions at 30 °C or pre-splicing control conditions at 0 °C. Incubation temperatures, 0 or 30 °C, are indicated above each lane. The identity of the pre-mRNA and of the mRNA spliced products is schematically drawn next to the autoradiogram. The nucleotide ladder is shown on the right. Retention of MRAP exon 3 (L1-MRAP-L2) is seen with MRAP wt whilst aberrant splicing resulting in skipping of MRAP exon 3 (L1–L2) is detected in MRAP mt. Bands corresponding to correct and aberrant mRNA splice products are indicated by the arrows. Full colour version of this figure available via http://dx.doi.org/10.1530/EJE-11-0581.