Abstract
Fibrillin-1 and fibrillin-2 are structural components of the extracellular matrix which are also involved in modulating local TGFβ and BMP bioavailability. Loss of fibrillin-1 or fibrillin-2 is associated with perturbed osteoblast maturation principally as the result of unbalanced TGFβ and BMP signaling. Here, we demonstrated that stable expression of small hairpin RNAs against fibrillin-1 (Fbn1) or fibrillin-2 (Fbn2) transcripts in the clonal osteoprogenitor cell line Kusa-A1 led to the same phenotypic and molecular manifestations as germline Fbn1- or Fbn2-null mutations in primary calvarial osteoblast cultures. Proof-of-concept experiments are also presented showing that Fbn1- or Fbn2-silenced Kusa-A1 cell lines are suitable models to identify candidate determinants of osteogenesis which are under the control of extracellular microfibrils. Specific findings included: the inference of a potential role for fibrillin-1-mediated cell–matrix interactions in regulating Kusa-A1 proliferation; the possibility of fibrillin-2 involvement in modulating the activity of transcription factor Runx2 by restricting microRNA expression and/or processing; and the suggestion that fibrillin-1 and fibrillin-2 influence Notch signaling indirectly by differentially regulating BMP signaling. Collectively, the data reiterated the notion that fibrillin-1 and fibrillin-2 exert opposite effects on osteoblast differentiation through the discrete modulation of a broad network of interacting signaling molecules.
Keywords: BMP, Fibrillin, Notch, Osteoblast, differentiation, Runx2, TGFβ, Cell culture
Introduction
TGFβ family members are potent determinants of multiple developmental programs and physiological processes, including bone formation and remodeling (Alliston et al. 2008). TGFβs promote the commitment of osteoblast progenitors and inhibit their maturation, while BMPs cooperate in the former process and induce the latter one (Alliston et al. 2008). A poorly understood mechanism of TGFβ family regulation is how binding to components of the extracellular matrix (ECM) influences the distribution, presentation and/or activation of the ligands (Ramirez and Rifkin 2009). Recent studies of microfibril-deficient mice have implicated fibrillin-1 and fibrillin-2 in bone formation through the differential control of local TFGβ and BMP signals during osteoblast maturation (Nistala et al. 2010). Fibrillin-1 and fibrillin-2 are ubiquitous glycoproteins that self-polymerize into filamentous microfibrils which guide elastic fiber formation and bind latent TGFβ-binding proteins (LTBPs) and BMP pro-peptides (Ramirez and Rifkin 2009). Primary calvarial osteoblasts (cOb) deficient for fibrillin-2 fail to mature due to inappropriately high TGFβ activity, whereas cOb deficient for fibrillin-1 mature more rapidly than the wild-type counterparts because TGFβ and BMP signaling are both elevated (Nistala et al. 2010).
Albeit informative, the study of primary cOb is, however, limited by the significant heterogeneity of the cell cultures, which restricts the yield of mature osteoblasts and often obscures the identification of critical stage-specific events, and by the need for pooling together data from multiple cell preparations, which is time-consuming and costly as well as subject to phenotypic and/or experimental variability. An important finding of previous investigations is that wild-type cOb transiently expressing small interfering (si) RNAs against Fbn1 or Fbn2 transcripts replicate the maturation defects of cOb harboring germline Fbn1- or Fbn2-null mutations (Nistala et al. 2010). The present study exploited this observation with two main goals in mind. The first goal was to compare the phenotypes of clonal osteoprogenitor (OP) cell lines stably expressing small hairpin (sh)RNA against Fbn1 or Fbn2 transcripts (shFbn1OP and shFbn2OP cell lines, respectively) to those of Fbn1- or Fbn2-null cOb cultures. The second goal was to gather proof-of-concept evidence in support of the notion that the clonal nature of the Fbn-silenced OP lines makes them attractive models to interrogate the functional role of fibrillin microfibrils through different stages of osteogenic differentiation. The results demonstrated phenotypic equivalence between shFbnOP cell lines and primary cOb cultures from Fbn-null mice, in addition to identifying new molecular abnormalities secondary to perturbations of TGFβ family bioavailability and/or cell-matrix interactions.
Materials and methods
Cell cultures
Kusa-A1 cells (RIKEN BRC Cell Bank) were infected with MISSION® shRNA Control Lentiviral Transduction Particles-SHC002V (shSrcOP cells) or fibrillin-specific MISSION® shRNA Lentiviral Transduction Particles (shFbnOP cells) (Sigma-Aldrich) (Smaldone et al. 2011). Lentivirus-infected Kusa-A1cells were maintained at 37°C in a sterile and humidified atmosphere of 5% CO2 and grown in α-MEM culture medium containing 10% fetal bovine serum (FBS; Atlanta Biologicals) and supplemented with puromycin (2 ng/ml) to select cells stably expressing the various shRNA. For differentiation assay, cells were grown to confluency (day 0) and then induced to differentiate by supplementing the culture medium with 50 µg/ml ascorbic acid and 10 mM β–glycerophosphate; mineral deposits were visualized by alizarin red staining. In some experiments cells were treated with 100 ng/ml of BMP2 (R&D Systems) or 100 ng/ml of Noggin (R&D Systems). Cells were only used at early passages (passages 3–7) to minimize potential problems of phenotypic drift.
Cell transfections
Cells were transiently co-transfected using Lipofectamine 2000 (Invitrogen) with 5 ng of the control plasmid TK-Renilla-Luc (Promega) and 500 ng of reporter plasmids for Runx2 (6xOSE2-OC-Luc) or Osx (pGL3Osx-Luc) (generous gifts of Drs. de Crombrugghe and Chen). Luciferase assays were performed on 3 independent samples (each in duplicate) and evaluated as previously described (Carta et al. 2009; Nistala et al. 2010; Smaldone et al. 2011).
Immunoblots
Total protein extracts were prepared from cell cultures and analyzed by Western blot hybridization as previously described (Carta et al. 2009; Nistala et al. 2010). Antibodies included those against mouse Smad2/3, pSmad2/3, pSmad1/5/8, PCNA (1:1,000 dilution; Cell Signaling), collagen I (1:1,000 dilution; Biogenesis) Runx2 (1:1,000 dilution; Abcam), the intracellular domain of Notch1 (NICD; Santa Cruz Biotechnologies) or β-actin (1:10,000 dilution; Sigma-Aldrich). Immunoblots were performed on 3 independent samples each run in duplicate.
RNA analyses
Total RNA for real-time quantitative PCR (q-PCR) was isolated from cultured cells and processed as previously detailed (Carta et al. 2009; Nistala et al. 2010; Smaldone et al. 2011). Amplification primers were purchased from SABiosciences; cycling conditions were 95°C for 10 min followed by 40 cycles consisting of 95°C for 15 s denaturation, 60°C for 30 s annealing and 72°C for 30 s extension. Comparative quantification was carried out using 3 independent samples each analyzed in triplicate.
Results and discussion
Fbn-silenced OP cell lines are phenotypically equivalent to germline Fbn-deficient cOb cells
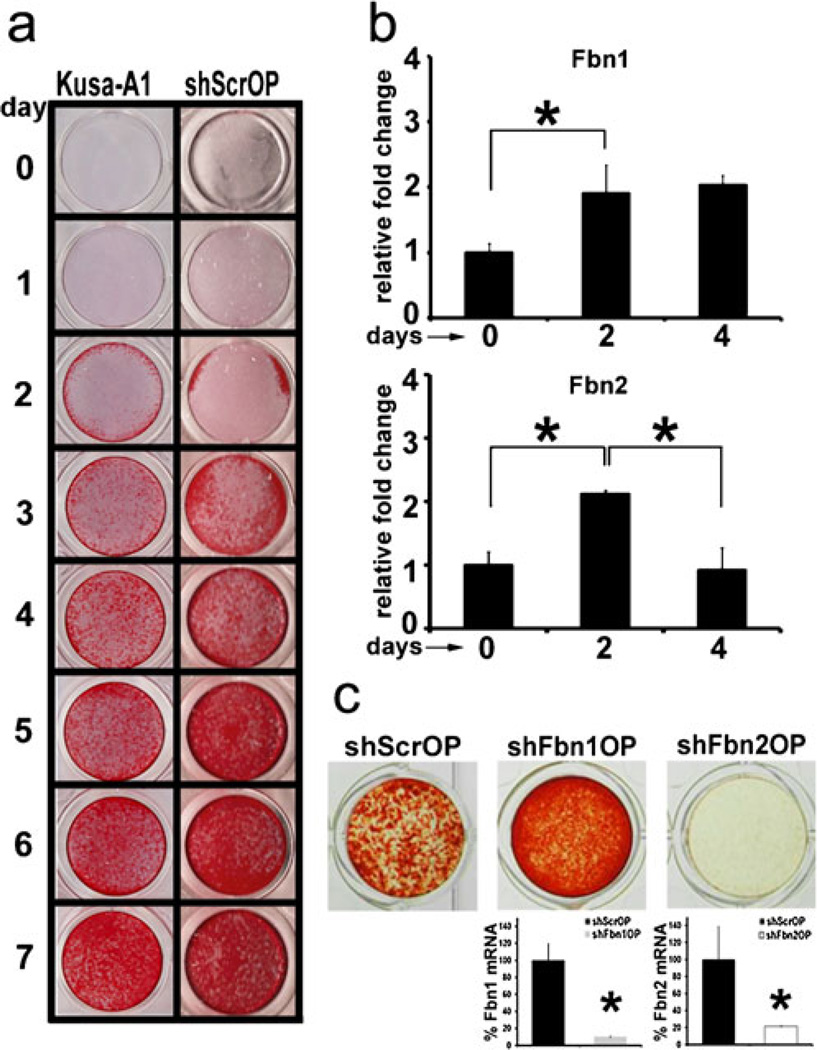
A screen of several osteogenic cell lines, including ST-2 and MC3T3 cells, that were infected with lentiviruses expressing scrambled shRNA (shSrc), shFbn1 or shFbn2, identified the OP clone line Kusa-A1 (Kawashima et al. 2005) as the one which differentiated into mature (mineralizing) osteoblasts more rapidly and more homogenously. Mineral (alizarin red-positive) deposits by control shScrOP cultures appeared 2 days after addition of osteoinductive supplements (OSs) and accumulated strongly and uniformly for the next 5 days in the same manner as in uninfected Kusa-A1 cell cultures (Fig. 1a). Parallel q-PCR analyses revealed a 2-fold increase in the amount of Fbn1 and Fbn2 transcripts in day 2 differentiating Kusa-A1 cells (Fig. 1b); in contrast, Fbn2 transcripts were noted to decline back to baseline 4 days after OS administration (Fig. 1b). Further evaluation of the q-PCR data estimated that day 2 differentiating Kusa-A1 cells contain 3.5 × 104 copies of Fbn1 transcripts and 2.26 × 102 copies of Fbn2 transcripts per 2.5 ng of input RNA. As a point of reference, the number of Col1a1 transcripts, which encode the major structural component of the bone matrix (Ramirez 2009), was calculated to be 2.25 × 106 copies per 2.5 ng of input RNA at the same stage of Kusa-A1 differentiation. Silencing of Fbn1 (shFbn1OP) or Fbn2 (shFbn2OP) expression in OS-treated Kusa-A1 cultures increased mineral deposition in the former and decreased it in the latter (Fig. 1c). Moreover, q-PCR analyses validated the specificity and extent of gene silencing in shFbn1OP and shFbn2OP cells (Fig. 1c). These results therefore equated the phenotypes of Fbn-null primary cOb and Fbn-silenced clonal OP cells in addition to reiterating the cell (culture)-autonomous nature of Fbn mutations (Nistala et al. 2010). Parenthetically, comparable phenotypes were observed in the other Fbn-silenced osteogenic cell lines we tested in the screen (data not shown).
Fig. 1.
Perturbed differentiation of shFbn1OP and shFbn2OP cells. a Alizarin red-positive calcium deposits in Kusa-A1 and shScrOP cell cultures before (day 0) and after (days 1–7) OS addition. b q-PCR quantification of Fbn1 and Fbn2 mRNAs in shScrOP cells at days 0, 2 and 4 of differentiation; control values are arbitrarily indicated as 1 unit. c Illustrative images of alizarin red-positive calcium deposits of shScrOP, shFbn1OP, and shFbn2OP cultures 4 days after OS addition. Histograms summarize the relative levels of Fbn1 or Fbn2 transcripts in control and silenced OP cells; control values are arbitrarily indicated as 100%. Asterisks in panels (b) and (c) indicate statistical significance (p<0.05)
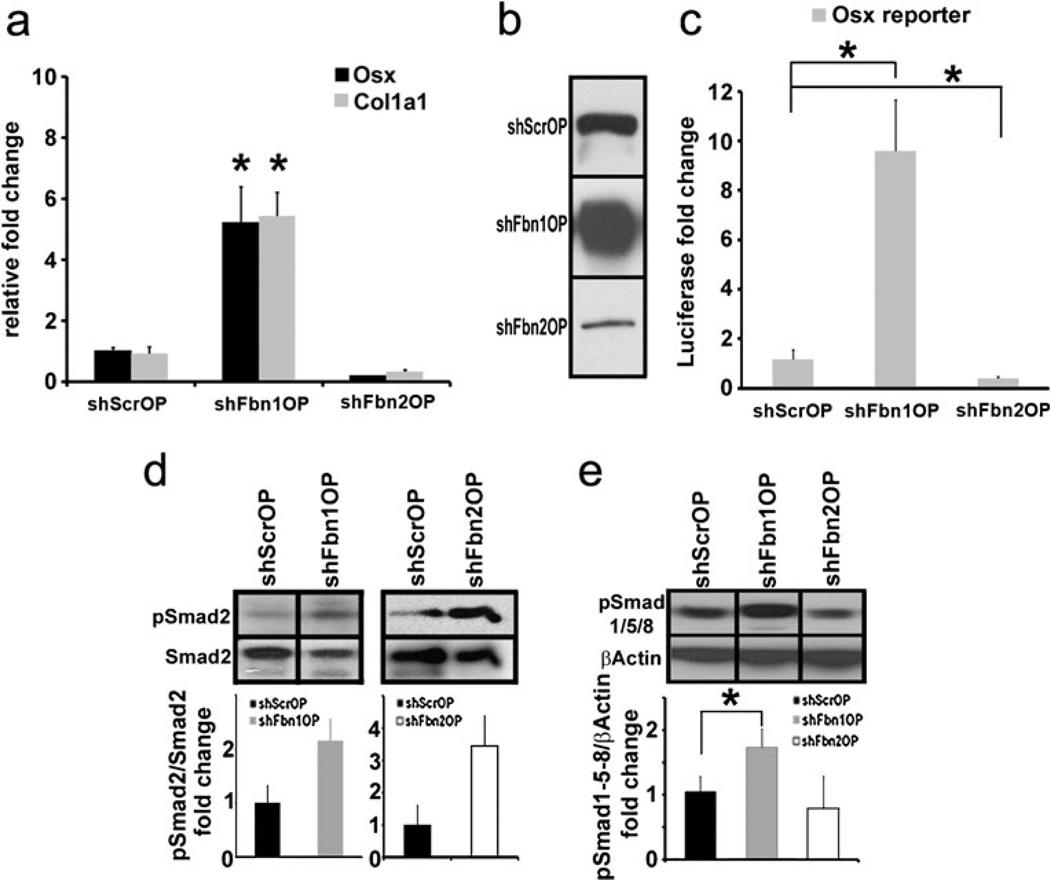
As per the previous study of fibrillin-deficient cOb cultures (Nistala et al. 2010), the above conclusion was corroborated and extended by monitoring key markers of osteogenic differentiation as well as TGFβ and BMP signaling in shFbn1OP and shFbn2OP cells 2 days after OS administration. Molecular markers included transcription factor Osterix (Osx), the major regulator of pre-osteoblast maturation (Nakashima et al. 2002), and collagen I, the structural template of bone matrix mineralization (Ramirez 2009). Similar to Fbn1- and Fbn2-null cOb cultures (Nistala et al. 2010), shFbn1OP and shFbn2OP cells, respectively, displayed substantial increases or decreases of both Osx and Col1a1 transcripts (Fig. 2a). Western blot analyses of collagen I proteins and cell transfection assays of an Osx-reporter plasmid further substantiated the q-PCR data (Fig. 2b, c). Like Fbn1- and Fbn2-null cOb cultures (Nistala et al. 2010), assessment of activated (phosphorylated) Smad2/3 and Smad1/5/8 levels indicated abnormally high TGFβ signaling in maturing shFbn1OP and shFbn2OP cultures and greater than normal BMP activity only in maturing shFbn1OP cultures (Fig. 2d, e). Together, cOb and OP culture experiments indicate that loss of fibrillin-1 or -2 differentially impact bone matrix maturation and mineralization in part as result of impaired Osx-driven collagen I production secondary to distinct perturbations in local TGFβ and BMP bioavailability (Nistala et al. 2010). Incidentally, the finding that Fbn2 (but not Fbn1) silencing promotes BMP signaling and expression of the osteoblast marker osteocalcin in C2C12 cells induced by low serum culture conditions to differentiate into myoblasts provided further evidence for the discrete and tissue-specific roles of fibrillins in controlling TGFβ and BMP bioavailability (data not shown).
Fig. 2.
Abnormal Osx and Col1a1 expression and TGFβ and BMP signaling in shFbnOP cells. a q-PCR quantification of Osterix (Osx) and collagen I (Col1a1) transcripts in shScrOP, shFbn1OP and shFbn2OP cells. b Collagen I immunoblots of protein extracts from shScrOP, shFbn1OP and shFbn2OP; the same amount of total proteins (25 µg) was loaded in each of the samples. c Osx promoter-driven luciferase activity in shScrOP, shFbn1OP and shFbn2OP cells expressed as average fold induction over control±SD. d pSmad2 immunoblots of protein extracts from shFbn1OP (left), shFbn2OP (right) and the control shScrOP cells lines with histograms summarizing the average pSmad2/Smad2 ratios±SD. e pSmad1/5/8 immunoblots of protein extracts from shScrOP cells and shFbn1OP or shFbn2OP cells with histograms summarizing the relative ratios of pSmad1/5/8 over β-actin±SD. Asterisks in relevant panels indicate statistical significance (p<0.05)
Differentiating Fbn-silenced OP lines exhibit distinct abnormalities in osteogenic determinants
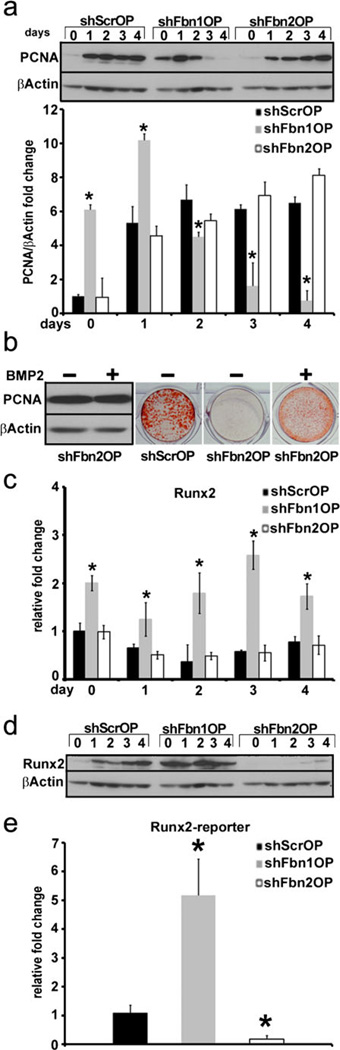
The second set of experiments compared three key aspects of osteogenic differentiation in shScrOP, shFbn1OP and shFbn2OP cultures, namely cell proliferation, Runx2 activity and Notch1 signaling. PCNA levels were monitored in the three OP lines from before OS administration (day 0) until advanced matrix mineralization (day 4). Control shScrOP cultures showed a rapid increase in PCNA protein accumulation following OS administration which plateaued at around day 2, immediately before completion of ECM maturation and appearance of mineral deposits (Fig. 3a). Differentiating shFbn2OP cells displayed virtually the same pattern as shScrOP cells (Fig. 3a). In contrast to the nearly undetectable PCNA levels in day 0 shScrOP and shFbn2OP cultures, the cell proliferation marker was already high in non-stimulated shFbn1OP cultures, increased further in response to OS administration peaking at day 1, and declined rapidly soon after reaching baseline levels by day 3 (Fig. 3a). These findings therefore implied that the rate of osteoblast proliferation depends more on the extracellular deposition of fibrillin-1 than fibrillin-2 microfibrils. Given that elevated TGFβ signaling is a common feature of shFbn1OP and shFbn2OP cultures (Fig. 2d), greater shFbn1OP proliferation could reflect the effect of either augmented BMP signaling in these mutant cells or cell-matrix interactions uniquely mediated by fibrillin-1 microfibrils. We favor the latter possibility because preliminary evidence showed that BMP2 treatment rescues maturation of shFbn2OP cells without significantly changing PCNA levels (Fig. 3b). Parenthetically, BMP2 treatment was previously shown to counteract the inhibitory action of promiscuous TGFβ signaling and thus improve the maturation defect of Fbn2-null cOb cultures (Nistala et al. 2010). We also speculate that the greater amount and steady production of fibrillin-1 compared to fibrillin-2 molecules (Fig. 1b) are probably responsible for fibrillin-1-mediated cell-matrix interactions that modulate the proliferation of progenitor and maturing osteoblast.
Fig. 3.
shFbnOP cells exhibit abnormal proliferation and Runx2. activity. a PCNA immunoblots of protein extracts from shScrOP, shFbn1OP, and shFbn2OP cells collected at the indicated time points; histograms summarize the ratios between PCNA and β-actin. b PCNA immunoblots of protein extracts from day 2 shFbn2OP cells cultured with or without BMP2 and illustrative examples of alizarin red-stained day 4 shFbn2OP cell cultured with or without BMP2. c q-PCR analysis of Runx2 mRNA levels in shScrOP, shFbn1OP and shFbn2OP cells collected at the indicated time points; average fold changes are reported±SD. d Runx2 and β-actin immunoblots of protein extracts from shScrOP, shFbn1OP and shFbn2OP cells lines collected at the indicated time points. e Runx2 promoter-driven luciferase activity in shScrOP, shFbn1OP and shFbn2OP cells expressed as average fold induction over control±SD. Asterisks in relevant panels indicate statistical significance (p<0.05)
Important differences were also noted with regard to transcription factor Runx2, the major determinant of OP commitment (Ducy et al. 1997). Differentiating shFbn1OP cells displayed higher than normal levels of Runx2 mRNA and protein (Fig. 3c, d), which translated into greater transcriptional activity of a transiently transfected Runx2-responsive plasmid (Fig. 3e). On the other hand, Runx2 protein levels and activity of a Runx2 reporter plasmid were substantially less in shFbn2OP than shSrcOP cells (Fig. 3d, e), even though the two cell lines displayed nearly identical profiles of Runx2 mRNA accumulation (Fig. 3c). These results imply that fibrillin-1 and fibrillin-2 are, respectively, involved in restricting and promoting Runx2 activity and that fibrillin-2 exerts its positive action through a yet to be defined post-transcriptional mechanism. One attractive possibility, which is supported by recent studies of osteogenic differentiation (Li et al. 2008, 2009; Huang et al. 2010; Gaur et al. 2010), is that fibrillin-2 may indirectly inhibit the expression and/or processing of microRNAs that target positive regulators of OP commitment and differentiation, such as Runx2.
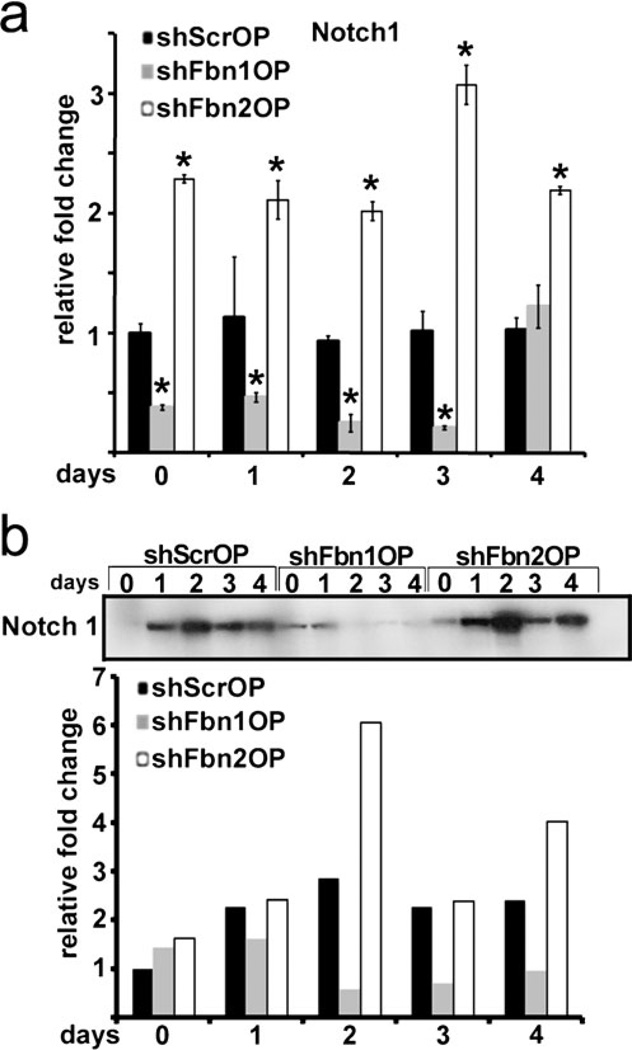
Lastly, Notch signaling was compared in the OP-silenced lines in view of its inhibitory action on the differentiation of Kusa-A1 mesenchymal cells as well as C2C12, ST-2 and MC3T3 cells (Nofziger et al. 1999; Sciaudone et al. 2003; Shindo et al. 2003; Deregowski et al. 2006). Consistent with their opposite patterns of cell differentiation (Fig. 1c), the shFbn1OP and shFbn2OP lines respectively displayed lower and higher than normal levels of Notch1 transcripts and the processed NICD peptide (Fig. 4a, b). As TGFβ signaling is elevated in both shFbn1OP and shFbn2OP cells (Fig. 2d), we next evaluated whether the opposite profiles of Notch1 expression may in part reflect the difference in BMP signaling between the two cell lines (Fig. 2e). Accordingly, Notch1 mRNA levels were assessed in control and Fbn-silenced cells cultured for 2 or 4 days under different experimental conditions, as these time points best illustrate the difference in Notch1 expression between shScrOP and shFbnOP cells (Fig. 2b). Addition of the BMP antagonist Noggin to shFbn1OP cultures, which are otherwise characterized by higher than normal BMP signaling (Fig. 2e), led to 1.7- to 2-fold increase in Notch1 mRNA levels compared to untreated mutant cells and, consequently, near normalization of the Notch1 mRNA ratio between control and Fbn1-silenced cells (Table 1). Conversely, addition of BMP2 to shFbn2OP cells, which otherwise display normal BMP signaling (Fig. 2e), resulted in ~2-fold less Notch1 mRNA levels compared to untreated cells and, consequently, reversion of the Notch1 mRNA ratio between control and Fbn2-silenced cells (Table 1). Taken at face value, these results suggest that abnormally high BMP signaling negatively impacts Notch1 expression in shFbn1OP cells. As constitutive Notch signaling in ST-2 cells indirectly opposes BMP2 stimulation of alkaline phosphatase activity (Deregowski et al. 2006), our finding also raised the intriguing possibility of a feed-back loop between the two signaling pathways. While ongoing investigations are testing this and related hypotheses, the present study is nonetheless the first to reveal an indirect role of the ECM in regulating Notch signaling during early osteoblast differentiation.
Fig. 4.
Opposite patterns of Notch signaling in shFbn1OP and shFbn2 OP cells. a Notch1 mRNA levels in shScrOP, shFbn1OP and shFbn2OP cells collected at the indicated time points. b NICD immunoblots of protein extracts from shScrOP, shFbn1OP, and shFbn2OP cells collected at the indicated time points
Table 1.
Notch1 mRNA ratio between shScrOP and shFbnOP cellsa
| untreated | Noggin | BMP2 | |
|---|---|---|---|
| shFbn1OP | Day 2 1.71±0.65 | Day 2 1.01±0.09 | NA |
| Day 4 2.03±0.91 | Day 4 0.96±0.36 | ||
| shFbn2OP | Day 2 0.79±0.47 | NA | Day 2 1.55±0.82 |
| Day 4 0.65±0.03 | Day 4 1.29±0.96 |
mRNA levels were normalized to β-actin; values represent the average of 3 independent assays each performed in triplicate
Summary
Here, we exploited the cell (culture)-autonomous nature of fibrillin-1 or fibrillin-2 deficiency to establish clonal shFbnOP lines that replicate the phenotypic and molecular features of primary Fbn1- or Fbn2-null cOb cultures. We also documented the value of these clonal cell lines to identify molecular abnormalities that are responsible for perturbed differentiation of pre-osteoblasts deficient for either fibrillin-1 or fibrillin-2 microfibrils. The work therefore provided the basis for ongoing studies aimed at elucidating the precise mechanisms whereby fibrillin-1 and fibrillin-2 exert opposite effects on OP cell proliferation, Runx2 activity, and Notch signaling.
Acknowledgements
The authors thank Drs. de Crombrugghe and Chen for reagents and Ms. Karen Johnson for organizing the manuscript. This work was supported by NIH grant AR42044.
References
- Alliston T, Piek E, Derynck R. TGF-β family signaling in skeletal development, maintenance and disease. In: Derynck R, Miyazono K, editors. The TGF-β family. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2008. pp. 667–723. [Google Scholar]
- Carta L, Smaldone S, Zilberberg L, Loh D, Dietz HC, Rifkin DB, Ramirez F. p38 MAPK is an early determinant of promiscuous Smad2/3 signaling in the aortas of fibrillin-1 (Fbn1)-null mice. J Biol Chem. 2009;284:5630–5636. doi: 10.1074/jbc.M806962200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deregowski V, Gazzerro E, Priest L, Rydziel S, Canalis E. Notch1 overexpression inhibits osteoblastogenesis by suppressing Wnt/β-catenin but not bone morphogenetic protein signaling. J Biol Chem. 2006;281:6203–6210. doi: 10.1074/jbc.M508370200. [DOI] [PubMed] [Google Scholar]
- Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. Osf2/ Cbfal: a transcriptional activator of osteoblast differentiation. Cell. 1997;89:747–754. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- Gaur T, Hussain S, Mudhassani R, Parulkar I, Colby JL, Frederick D, Kream BE, van Wijnen AJ, Stein JL, Stein GS, Jones SN, Lian JB. Dicer inactivation in osteoprogenitor cells compromises fetal survival and bone formation, while excision in differentiated osteoblasts increases bone mass in the adult mouse. Dev Biol. 2010;340:10–21. doi: 10.1016/j.ydbio.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Zhao L, Xing L, Chen D. MicroRNA-204 regulates Runx2 protein expression and mesenchymal progenitor cell differentiation. Stem Cells. 2010;28:357–364. doi: 10.1002/stem.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima N, Shindo K, Sakamoto K, Kondo H, Umezawa A, Kasugai S, Perbal B, Suda H, Takagi M, Katsube K. Molecular and cell biological properties of mouse osteogenic mesenchymal progenitor cells. Kusa J Bone Miner Metab. 2005;23:123–133. doi: 10.1007/s00774-004-0550-y. [DOI] [PubMed] [Google Scholar]
- Li Z, Hassan MQ, Volinia S, van Wijnen AJ, Stein JL, Croce CM, Lian JB, Stein GS. A microRNA signature for a BMP2-induced osteoblast lineage commitment program. Proc Natl Acad Sci USA. 2008;105:13906–13911. doi: 10.1073/pnas.0804438105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Hassan MQ, Jafferji M, Aqeilan RI, Garzon R, Croce CM, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Biological functions of miR-29b contribute to positive regulation of osteoblast differentiation. J Biol Chem. 2009;284:15676–15684. doi: 10.1074/jbc.M809787200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, de Crombrugghe B. The novel zinc finger-containing transcription factor Osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- Nistala H, Lee-Arteaga S, Smaldone S, Siciliano G, Ono R, Sengle G, Arteaga-Solis E, Levasseur R, Ducy P, Sakai LY, Karsenty G, Ramirez F. Fibrillin-1 and-2 differentially modulate endogenous TGFβ and BMP bioavailability during bone formation. J Cell Biol. 2010;190:1107–1121. doi: 10.1083/jcb.201003089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nofziger D, Miyamoto A, Lyons KM, Weinmaster G. Notch signaling imposes two distinct blocks in the differentiation of C2C12 myoblasts. Development. 1999;126:1689–1702. doi: 10.1242/dev.126.8.1689. [DOI] [PubMed] [Google Scholar]
- Ramirez F. The skeletal system. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2009. Extracellular matrix in the skeleton; pp. 341–353. [Google Scholar]
- Ramirez F, Rifkin DB. Extracellular microfibrils: contextual platforms for TGFβ and BMP signaling. Curr Opin Cell Biol. 2009;21:616–622. doi: 10.1016/j.ceb.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciaudone M, Gazzerro E, Priest L, Delaney AM, Canalis E. Notch1 impairs osteoblastic differentiation. Endocrinology. 2003;144:5631–5639. doi: 10.1210/en.2003-0463. [DOI] [PubMed] [Google Scholar]
- Shindo K, Kawashima N, Sakamoto K, Yamaguchi A, Umezawa A, Tagaci M, Katsube K, Suda H. Osteogenic differentiation of the mesenchymal progenitor cells Kusa is suppressed by Notch signaling. Exp Cell Res. 2003;290:370–380. doi: 10.1016/s0014-4827(03)00349-5. [DOI] [PubMed] [Google Scholar]
- Smaldone S, Olivieri J, Gusella GL, Moroncini G, Gabrielli A, Ramirez F. Ha-Ras stabilization mediates pro-fibrotic signals in dermal fibroblasts. Fibrogenesis Tissue Repair. 2011;4:8. doi: 10.1186/1755-1536-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]