FIG. 1.
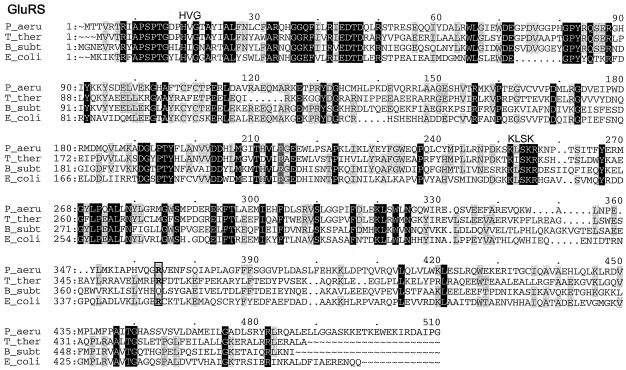
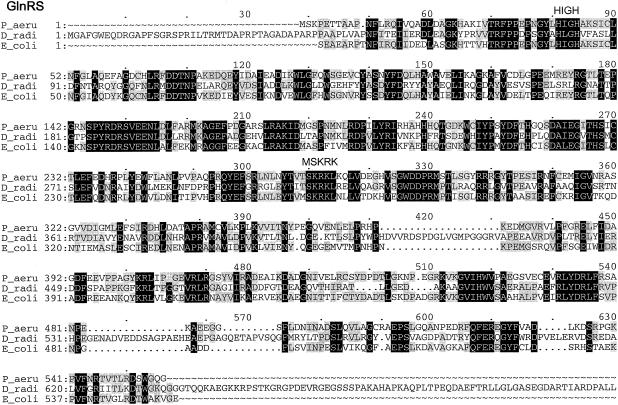
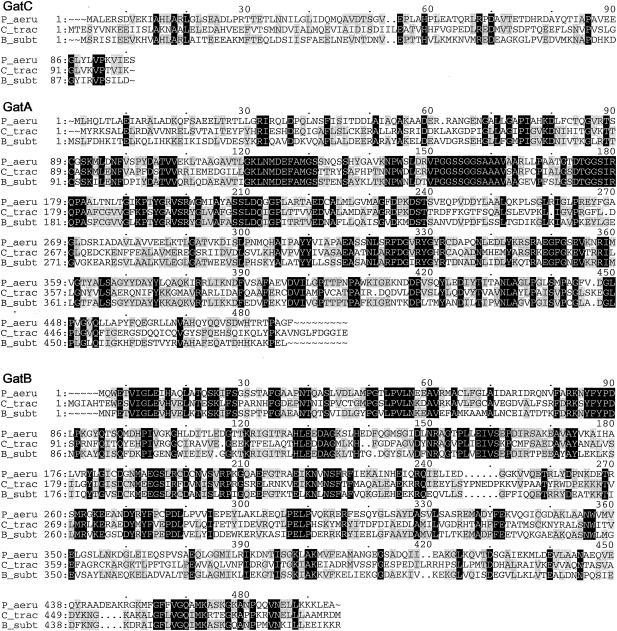
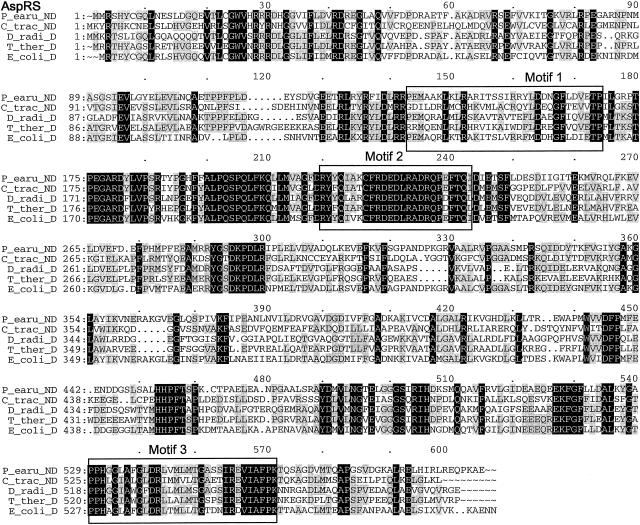
Multiple-sequence alignments of P. aeruginosa PAO1 GluRS, GlnRS, GatC, GatA, GatB, and AspRS with a few respective orthologs from other bacterial species, created by using the Pileup program. The organisms were as follows: P_aeru, P. aeruginosa PAO1; T_ther, T. thermophilus; B_subt, B. subtilis; E_coli, E. coli; D_radi, Deinococcus radiodurans; C_trac, C. trachomatis. The GluRS sequences used were as follows: P_aeru, PA3134; T_ther, P27000; B_subt, P222450; E_coli, P04805. The aligned residues Arg358 of T. thermophilus GluRS-D, Arg358 of P. aeruginosa PAO1, and GluRS and Gln358 of B. subtilis GluRS-ND are boxed. The GlnRS sequences used were as follows: P_aeru, Q9I2U8; D_radi, P56926; E_coli, BAA35328. The GatC, GatA, and GatB sequences used were, respectively, as follows: P_aeru, AAG07870, AAG07871, AAG07872; C_trac, NP_219504, NP_219505, NP_219506; B_subt, O06492, CAB12488, O30509. The AspRS sequences used were as follows: P_aeru, NP_249654; C_trac, O84546; D_radi_D, NP_295070; T_ther_D, P36419; E_coli_D, NP_288303. Residues which were identical in all sequences of each multiple alignment are printed in white on black, and those conserved in at least three of the four GluRS sequences, two of three sequences (GlnRS, GatC, GatA, and GatB), or three or four of the five AspRS sequences are shaded.