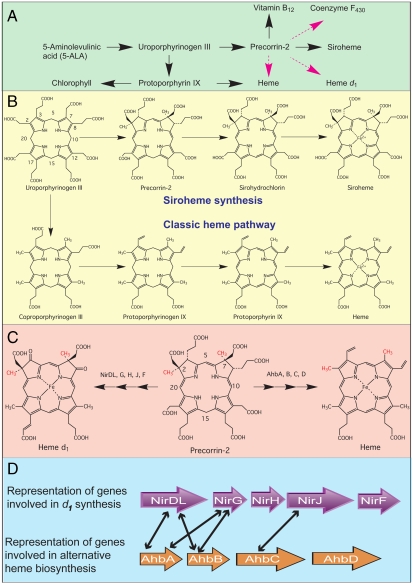
Fig. 1.
Pathways and genes of modified tetrapyrrole biosynthesis. (A) Outline syntheses of modified tetrapyrroles highlighting key intermediates along the branched pathway. Known reaction sequences are shown in black whereas those not yet elucidated are highlighted in magenta. (B) The known transformation of uroporphyrinogen III into siroheme and heme is shown. The numbering system for the tetrapyrrole macrocycle is highlighted on uroporphyrinogen III. (C) Precorrin-2 as the template for both d1 heme and heme synthesis is shown, with the latter derived via the alternative heme pathway. In both cases the two methyl groups at C2 and C7, highlighted in red, are derived from AdoMet. The enzymes involved in the transformation of precorrin-2 into d1 heme (Nir enzymes) and heme (Ahb enzymes) are shown. (D) The similarity between some of the Nir proteins implicated in d1 synthesis and proteins found in sulfate-reducing bacteria and Archaea thought to be associated with the Ahb pathway are indicated by black arrows.