Figure 1.
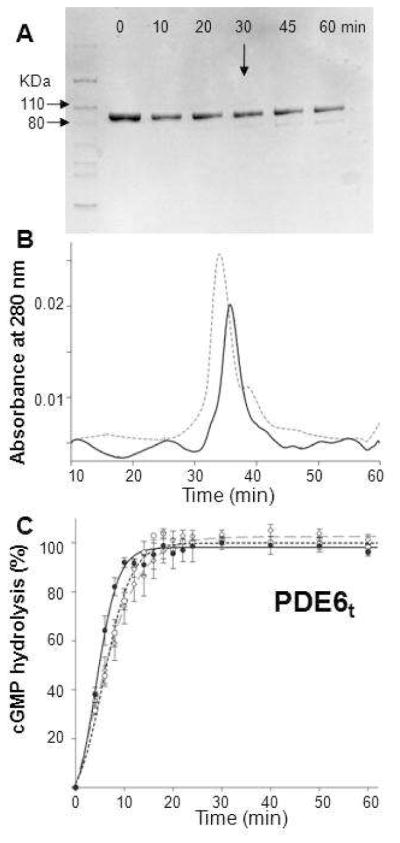
Preparation of trypsinized PDE6. A. The time course of trypsin digestion monitored by SDS–PAGE is shown. Purified PDE6 was mixed with trypsin at 100:1 (weight ratio) and incubated at room temperature for 10, 20, 30, 45 and 60 min. Protein without trypsin was included as a control (Lane 0). At 30 min (arrow), a weak band with a smaller molecular weight appeared. B. Size exclusion chromatography profile of PDE6t (30 min, room temperature). PDE6t (solid line) eluted at nearly the same position as untreated PDE6 (dashed line). C. PDE6t activity with cGMP (2 mM) as substrate. Enzyme activities monitored with the pH–sensitive fluorescent dye SNARF–1were positively correlated with increasing fluorescence intensity over time. PDE6t was generated as follows: Purified PDE6 was first mixed with trypsin at a 100:1 weight ratio for 10 min. Soybean trypsin inhibitor was then added at a 1:10 mass ratio to stop the reaction (dashed line, –○–). Alternatively, PDE6 was mixed with trypsin–agarose at 30 μg protein per 1 μl agarose for 30 min and the reaction was stopped by passing the sample through a filter to remove the trypsin–agarose. Trypsinized peptides then were removed either by gel filtration (dashed line, –◇–) or buffer exchange with a 50 kDa MWCO filter (Millipore) (solid line, –•–). Activities for PDE6t prepared by all three methods were identical.
