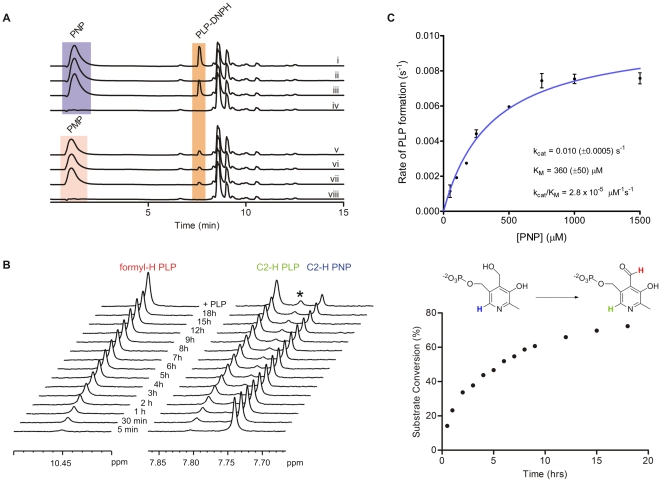
Figure 3. Reverse-phase HPLC, NMR, and spectrophotometric analysis of the PNPOx activity of Rv2607.
(A) HPLC chromatograms (270 nm) of reaction mixtures containing PNP (i-iv) or PMP (v-viii), FMN, and Rv2607, and the associated control reactions. All reactions were carried out in 25 mM potassium phosphate buffer (pH 7.8), incubated at 25°C for 3 h, and quenched with DNPH (0.7 mM final concentration). Where present, the reaction components were at the following concentrations: 1 mM PNP or PMP, 10 µM FMN, and 10 µM Rv2607. Reaction mixtures contained: (i) PNP, FMN, and Rv2607, (ii) PNP and FMN (enzyme negative control), (iii) PNP and Rv2607 (no added FMN), (iv) FMN and Rv2607 (substrate negative control), (v) PMP, FMN, and Rv2607, (vi) PMP and FMN (enzyme negative control), (vii) PMP and Rv2607 (no added FMN), (viii) FMN and Rv2607 (substrate negative control). The peak to the right of PLP-DNPH is DNPH, which has a retention time of 8.6 min. (B) 1H NMR analysis of the conversion of PNP into PLP with time. The enzymatic reaction (1 mM PNP, 28 µM enzyme, 10% D2O in 25 mM potassium phosphate buffer, pH 7.8) was incubated for 18 hours at 298 K in the spectrometer. Left; stacked 1H NMR spectra recorded at various time points and after the addition of authentic PLP. The starred peak corresponds to PLP hydrate. Right; a plot of percent substrate conversion versus time. Substrate conversion was determined by comparing integrals of the C2-1H signals associated with PLP (7.80 ppm) to that of PNP (7.75 ppm). (C) Michaelis-Menten plot for Rv2607 with PNP as a substrate. The rate of PLP formation was monitored spectrophotometrically (λmax = 388 nm, ε = 4900 cm−1M−1) for various concentrations of PNP. All solutions were made in 100 mM potassium phosphate buffer (pH 7.8).