Table 3. Estimated type I error probability for test of deviation from HWP of SNP2, a causal SNP to primary disease but unassociated with secondary phenotype (MAF = 40%), at 0.05 and 0.0001 significance levels in simulation studies* using different approaches for HWP testing.
| Approaches |

|
α = 0.05 | α = 0.0001 | ||||||

|

|
||||||||
| 0.1 | 0.3 | 0.5 | 0.7 | 0.1 | 0.3 | 0.5 | 0.7 | ||
| LRT_t | 0.1 | 0.627260 | 0.573700 | 0.550220 | 0.559390 | 0.053796 | 0.040312 | 0.034674 | 0.037743 |
| 0.3 | 0.210050 | 0.198150 | 0.200780 | 0.196990 | 0.003259 | 0.002515 | 0.002577 | 0.002789 | |
| 0.5 | 0.050007 | 0.049852 | 0.049675 | 0.051325 | 0.000071 | 0.000113 | 0.000062 | 0.000160 | |
| 0.7 | 0.216220 | 0.219080 | 0.217210 | 0.215410 | 0.003184 | 0.003390 | 0.003172 | 0.002850 | |
| mHWP_t | 0.1 | 0.631460 | 0.584970 | 0.536990 | 0.488360 | 0.053770 | 0.026639 | 0.010651 | 0.007235 |
| 0.3 | 0.210800 | 0.208130 | 0.200800 | 0.168160 | 0.003330 | 0.001737 | 0.000933 | 0.000374 | |
| 0.5 | 0.050602 | 0.054912 | 0.054477 | 0.048824 | 0.000073 | 0.000057 | 0.000016 | 0.000005 | |
| 0.7 | 0.218030 | 0.229660 | 0.232740 | 0.228650 | 0.003316 | 0.003497 | 0.002618 | 0.001366 | |
| LRT_d | 0.1 | 0.050558 | 0.050690 | 0.050830 | 0.051459 | 0.000092 | 0.000117 | 0.000148 | 0.000103 |
| 0.3 | 0.051329 | 0.055468 | 0.056119 | 0.052864 | 0.000089 | 0.000129 | 0.000150 | 0.000111 | |
| 0.5 | 0.050079 | 0.054422 | 0.055539 | 0.053985 | 0.000067 | 0.000120 | 0.000057 | 0.000145 | |
| 0.7 | 0.050483 | 0.051177 | 0.051220 | 0.050447 | 0.000094 | 0.000136 | 0.000103 | 0.000107 | |
| mHWP_d | 0.1 | 0.054121 | 0.054870 | 0.054884 | 0.055146 | 0.000067 | 0.000117 | 0.000097 | 0.000100 |
| 0.3 | 0.056680 | 0.061018 | 0.061881 | 0.057905 | 0.000077 | 0.000068 | 0.000089 | 0.000040 | |
| 0.5 | 0.048408 | 0.052943 | 0.053900 | 0.052227 | 0.000067 | 0.000145 | 0.000057 | 0.000132 | |
| 0.7 | 0.055468 | 0.056702 | 0.056614 | 0.056278 | 0.000024 | 0.000100 | 0.000055 | 0.000081 | |
| eLRT | 0.1 | 0.050108 | 0.049843 | 0.049292 | 0.050494 | 0.000096 | 0.000125 | 0.000095 | 0.000046 |
| 0.3 | 0.050273 | 0.050039 | 0.049661 | 0.050889 | 0.000065 | 0.000133 | 0.000101 | 0.000102 | |
| 0.5 | 0.049889 | 0.050504 | 0.049880 | 0.050395 | 0.000076 | 0.000065 | 0.000070 | 0.000105 | |
| 0.7 | 0.049888 | 0.049224 | 0.051004 | 0.050025 | 0.000068 | 0.000139 | 0.000087 | 0.000115 | |
| emHWP | 0.1 | 0.055240 | 0.050562 | 0.038818 | 0.025086 | 0.000070 | 0.000032 | 0.000012 | 0.000012 |
| 0.3 | 0.053956 | 0.046384 | 0.034750 | 0.022099 | 0.000047 | 0.000008 | 0.000012 | 0.000008 | |
| 0.5 | 0.052390 | 0.055258 | 0.046097 | 0.032279 | 0.000076 | 0.000041 | 0.000020 | <0.000001 | |
| 0.7 | 0.055004 | 0.054376 | 0.055794 | 0.054795 | 0.000002 | 0.000085 | 0.000046 | 0.000102 | |
*Simulation studies were based on 1,000,000 replicates, each replicate with 2,000 cases in terms of primary disease and 2,000 controls frequency-matched on secondary phenotype.
MAF: minor allele frequency.
LRT_t: LRT approach, using presence and absence of secondary phenotype as cases and controls.
mHWP_t: mHWP exact test, using presence and absence of secondary phenotype as cases and controls.
LRT_d: LRT approach, using presence and absence of primary disease as cases and controls.
mHWP_d: mHWP exact test, using presence and absence of primary disease as cases and controls.
eLRT: extended LRT approach.
emHWP: extended mHWP exact test.
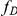 : prevalence of primary disease in general population.
: prevalence of primary disease in general population.
 : prevalence of secondary phenotype in general population.
: prevalence of secondary phenotype in general population.
