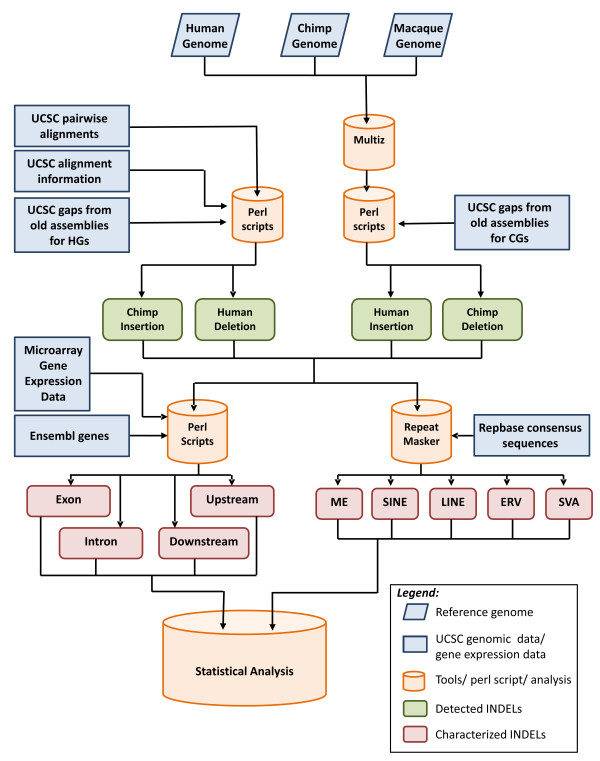
Figure 1.
Computational pipeline for the detection and characterization of human and chimpanzee insertions and deletions. Using information from the designated databases, we characterized insertions and deletions (INDELs) and analyzed them using various in-house Perl scripts and open source algorithms (Multiz, RepeatMasker [44] and Tandem Repeats Finder [45]). The multiple alignment program Multiz was used to classify chimpanzee gaps (CGs) as insertions or deletions. The UCSC Genome Browser [40] pairwise alignment databases were used for human gap (HG) classification as insertions or deletions. Human and chimpanzee INDELs were associated with the known human and chimpanzee Ensembl genes [30] obtained from the UCSC Table Browser (http://genome.ucsc.edu/cgi-bin/hgTables), and the presence of INDELs was correlated with the microarray gene expression data. INDEL sequences that were obtained from their corresponding reference genomes were searched for various repeat elements using RepeatMasker and Tandem Repeats Finder and classified according to the families of repeat sequences (partial or complete) present within each INDEL. The characterized INDELs were then assessed using various statistical analytical methods.